Patient characteristics
From January to December 2023, 30,243 SARS-CoV-2 tests were performed in paediatric subjects 0–17 years of age at the Bambino Gesù Children Hospital IRCCS in Rome, of which 1,510 (5.0%) resulted positive. Among them, 576 nasopharyngeal swabs were characterized by cycle threshold (Ct) < 30 or Antigenic Cut-off index (COI) > 100 and had demographic and clinical information available and were available for sequencing. In comparisons of the general and the identified SARS-CoV-2 infected population selected for potential sequencing, no differences were observed in demographic characteristics or clinical findings, except for mean cycle thresholds (p = 0.002) and COI values (p < 0.001), which represented the inclusion criteria for sequencing (Supplementary Table 1). Among the 576 selected patients, sequencing was successfully obtained for 458 (79.5%) patients. Demographic and clinical characteristics of the final 458 patients are reported in Table 1. Two hundred forty-nine (54.4%) were male. Median age was 0.6 (interquartile range [IQR] 0.3–1.4) years. Three hundred and thirty-two (72.5%) patients were < 1 year of age. Most individuals were Italian (N = 404, 88.2%) and lived in Lazio region (i.e. residing in the same region as the hospital, N = 415, 90.6%). One hundred and five (22.9%) patients presented with at least one comorbidity. The most prevalent comorbidities were being immunocompromised (22/105, 21.0%), cardiovascular disorders (15/105, 14.3%), genetic disorders (10/105, 9.5%), and neurological disorders (7/105, 6.7%). Among the other comorbidities, the following could be found: Asthma (N = 3), Bilateral Atresia Auris and Right Cryptorchidism (N = 1), Congenital left clubfoot (N = 1), Developmental Delay (N = 1), Ectopic Kidney (N = 1), Esophageal atresia (N = 1), Gastrointestinal diseases (N = 2), Multicystic Kidney (N = 1), Periodic Fever, Aphthous Stomatitis, Pharyngitis, Adenitis Syndrome (PFAPA, N = 1), Reactive airway disease (N = 2), Right Parietal Fracture (N = 1), Speech and Motor Delay, Microcephaly, and Facial Dysmorphisms (N = 1), Spina Bifida (N = 1), Tetraparesis, Dysphagia and Psychomotor Retardation Outcomes of Anoxic Damage from Methaemoglobinemia (N = 1), and Others Not Specified (N = 46).
Focusing on hospitalization, most patients presented to the emergency department (n = 443, 96.7%), 99 (21.6%) required hospitalization with a median (IQR) length of 4 (3–9) days, and 7 (1.5%) were admitted to the Intensive Care Unit (ICU). A higher incidence of lower respiratory tract infections (15.2% vs. 1.7%, p < 0.001) and gastrointestinal symptoms (10.1% vs. 2.87%, p = 0.004) were observed in hospitalized patients, whereas upper respiratory tract infections were more prevalent among non-hospitalized patients (83.3% vs. 39.4%, p < 0.001) (Supplementary Fig. 1). Furthermore, a significant proportion of hospitalized patients were asymptomatic (35.4%vs. 12.3%, p < 0.001) (Supplementary Fig. 1), likely due to comorbidities.
Table 1 Demographic and clinical characteristics of the 458 SARS-CoV-2-infected patients.Distribution of SARS-CoV-2 lineages and integrin binding domain
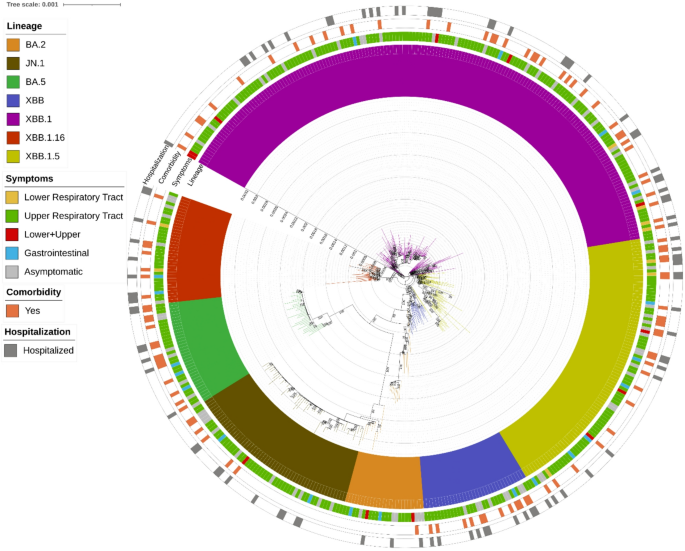
Most SARS-CoV-2 infections (75.0%) were caused by XBB and its sub-lineages, followed by JN.1 (12.4%), BA.5 (7.4%), and BA.2 (5.2%) (Fig. 1). Eleven different sub-lineages of interest were identified. The most prevalent were EG.5.1 (n = 77), JN.1 (n = 57), XBB.1.5 (n = 37), XBB.1.16 (n = 30), BQ.1 (n = 28), XBB.1.9 (n = 12) and XBB.2.3 (n = 10). All sub-lineages identified are reported in the Supplementary material.
Several specific integrin-binding motifs were present in the Omicron viral sequences. Among the 458 sequences analysed, only 3 presented the RGD (arginine/glycine/aspartic acid) motif, while most presented the RGN (arginine/glycine/asparagine) motif (Supplementary material). Additionally, the KGN (lysine/glycine/asparagine) and KGD (lysine/glycine/aspartic acid, already observed in SARS-CoV-1) motifs were present in 65 and 1 sequences, respectively (Supplementary material).
Clinical manifestations
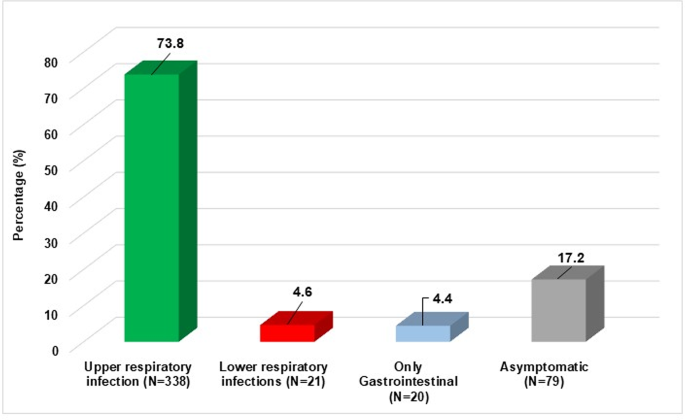
The majority of patients presented with upper respiratory infections (n = 338, 73.8%), followed by asymptomatic infections (n = 79, 17.2%), while a smaller proportion exhibited lower respiratory infections (n = 21, 4.6%) (Figs. 1 and 2).
By analysing the distribution of lineages in these infections we observed that lower infections were mainly characterised by the XBB.1 lineage (11/21, 52.4%), followed by the XBB.1.5 lineage (19/21, 19%) and then by the others with a frequency of less than 10%, except for BA.5, which was never observed. Even in upper infections, the main lineage involved was XBB.1 (145/338, 42.9%), followed by the lineage XBB.1.5 (16.6%) and JN.1 (13.3%). The lineage distribution in patients with asymptomatic infection was equally distributed (within 10%), except for XBB.1, which was found in 35.4%.
Estimated Maximum likelihood phylogenetic tree of the 458 SARS-CoV-2 sequences obtained from population aged ≤ 18 years. The maximum likelihood was inferred from a core genome alignment of 29,164 bp. The phylogeny was estimated with IQTREE using the best-fit model of nucleotide substitution GTR + F + R3 with 1,000 replicates and fast bootstrapping. The numbers on leaves represent the sample IDs, and bootstrap values higher than 90 are shown on branches. SARS-CoV-2 genomes were highlighted in different colours against omicron lineages (first, inner circle). Information regarding symptoms (second circle), comorbidities (third circle) and hospitalization (fourth, outer circle) were also reported. Among symptoms, lower and upper respiratory tract infections were considered regardless of gastrointestinal symptoms.
Distribution of clinical manifestation. A total of 458 infected patients are reported, including N = 358 (73.8%) presenting with upper respiratory infections, N = 21 (4.6%) with lower respiratory infections, N = 20 (4.4%) with gastrointestinal infections and N = 79 (17.2%) asymptomatic.
Correlation with hospitalization
Univariable and multivariable logistic regression models were used to assess whether the risk of hospitalization was associated with certain SARS-CoV-2 sub-lineages, clinical manifestations, or demographic characteristics (Table 2). The results showed that the presence of at least one comorbidity (versus no comorbidity) and involvement of the lower respiratory airways compared with asymptomatic patients were positively associated with hospitalization (adjusted OR [95% CI]: 5.59 [3.22–9.71], p-value < 0.001; and 3.16 [1.02–9.81], p-value = 0.046, respectively), while a negative association was observed with upper respiratory airways with respect to asymptomatic patients (0.25 [0.13–0.46], p-value < 0.001). These results were confirmed by performing the analysis under two extreme assumptions: treating all missing data as vaccinated and treating all missing data as unvaccinated (Supplementary Table 2).
In particular, the risk of hospitalization was significantly higher among patients with lower respiratory infections compared to those without (71.0% vs. 19.2%, p = 0.026), and ICU admissions were more frequent among patients with lower respiratory infections compare to those without (23.8% vs. 0.5%, p < 0.001). No associations were observed between the risk of hospitalization and specific Omicron lineages.
Table 2 Multivariable logistic regression analysis of factors associated with hospitalization.Viral and bacterial co-infections
Among the 21 patients with lower respiratory infection, nine (42.9%) presented with co-infection, namely four viral co-infections (1 Metapneumovirus, 1 Respiratory Syncytial Virus, 1 Rhinovirus A/B/C, and 1 Bocavirus 1/2/3 + Respiratory Syncytial Virus + Rhinovirus A/B/C), four bacterial co-infections (1 Pseudomonas aeruginosa + Staphylococcus aureus, 1 Staphylococcus aureus + Pseudomonas aeruginosa, 1 Escherichia coli, 1 Pseudomonas aeruginosa + Klebsiella pneumoniae + Enterococcus faecalis), and one viral and bacterial co-infection (Human Herpesvirus 6, HHV-6 + Staphylococcus aureus + Enterococcus faecalis, + Streptococcus pneumoniae).
Most co-infections primarily involved the respiratory tract, with eight patients showing respiratory involvement and pathogens isolated from respiratory samples including Metapneumovirus, RSV, Rhinovirus, Bocavirus, Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae, Enterococcus faecalis, Streptococcus pneumoniae, and Escherichia coli. However, in several cases, other body districts were also affected: blood co-infections were documented in two patients, one positive for Pseudomonas aeruginosa and the other for Staphylococcus aureus, Streptococcus pneumoniae, and HHV-6; gastrointestinal co-infections were identified in three patients through positive coprocultures or fecal PCR for Klebsiella pneumoniae, Enterococcus faecalis, and enteroaggregative Escherichia coli; additionally, urinary tract involvement was noted in one patient with a positive urinary antigen test for Streptococcus pneumoniae. Among these nine patients with co-infections, five were admitted to the ICU and presented with acute respiratory failure (Supplementary Table 3).
Among these 9 patients with co-infections, 5 were admitted to ICU and presented acute respiratory failure.