This approach not only offers a faster and more accurate way to measure plant traits in the field but also provides crucial insights into the molecular regulation of cotton architecture.
Cotton (Gossypium spp.), a key fiber crop for the global textile industry, relies heavily on plant architecture traits—such as height, branching, and stem growth—for yield and harvest efficiency. Upland cotton (G. hirsutum), which accounts for over 90% of global cotton production, particularly benefits from optimized plant height that allows higher planting density and smoother machine harvesting. Past agricultural revolutions, such as the introduction of dwarf genes into staple crops, have shown the profound impact of modifying height-related traits. However, traditional breeding for plant height requires slow and labor-intensive fieldwork. With rapid advances in genomics, identifying the genes that govern cotton height has become a research priority. Yet a major challenge remains: capturing accurate, high-throughput field data on plant height to match the scale of genetic datasets.
A study (DOI: 10.1016/j.plaphe.2025.100026) published in Plant Phenomics on 5 March 2025 by Zuoren Yang’s team, Chinese Academy of Agricultural Sciences, demonstrates the power of UAV-based multisensor platforms to overcome the bottleneck of field phenotyping.
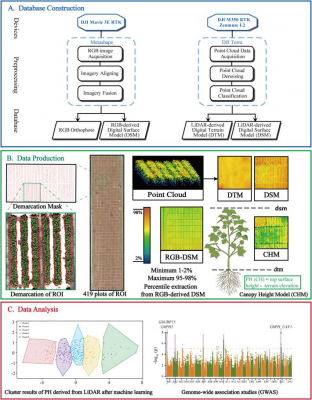
In this study, researchers employed UAV-mounted RGB cameras and LiDAR sensors to phenotype 419 cotton accessions across five developmental stages, extracting plant height (PH) values through linear regression and machine learning approaches before validating results against manual measurements. Linear regression analysis revealed that second-degree polynomial regression offered the strongest fit, yielding R² = 0.8974 for RGB-derived PH and R² = 0.9173 for LiDAR-derived PH compared with ruler-based measurements, with low error rates. When tested on all 419 accessions at later growth stages, correlations between UAV predictions and manual data remained robust, particularly for LiDAR (R = 0.89 and 0.87 at two time points). To refine prediction accuracy, 12 machine learning models were evaluated, where least angle regression for RGB and Huber regression for LiDAR delivered the best performance, achieving R² values of 0.914 and 0.934, respectively, with strong correlations to field measurements. Building on these reliable estimates, the team conducted K-means clustering of PH dynamics, dividing the cotton population into five distinct growth patterns, each with unique trajectories of height increase and peak timing. Subsequently, GWAS analysis using UAV-derived PH predictions identified 1,465 SNPs associated with plant height, including five consistent genomic signals across chromosomes A01, A02, A07, A10, and D11. Within these intervals, 34 annotated genes were detected, notably including GhPH1 and GhUBP15, both previously known regulators of cotton height, thereby confirming the accuracy of the UAV phenotyping platform. Importantly, a novel gene, GhPH_UAV1, was discovered, showing significantly reduced expression in a dwarf mutant compared with wild-type cotton. Functional validation demonstrated that overexpression of GhPH_UAV1 increased plant height by 16.48%, while CRISPR-Cas9 knockout reduced height by 21.44%, with corresponding changes in stem cell length. Together, these results demonstrate that UAV-based phenotyping, coupled with advanced data analysis, offers a powerful, scalable tool for dissecting the genetic regulation of complex agronomic traits.
By generating rapid, cost-effective, and accurate time-series data, researchers can better link genetic variation to crop performance under real-world conditions. For cotton breeding, the identification of GhPH_UAV1 offers a new genetic target to fine-tune plant architecture for higher yield, lodging resistance, and compatibility with dense planting and mechanical harvesters. More broadly, this strategy could be applied across crops such as maize, wheat, and rice, where plant height remains a cornerstone trait for agricultural productivity.
###
References
DOI
Original URL
https://doi.org/10.1016/j.plaphe.2025.100026
Funding information
This work was supported by the National Key R&D Program of China (2022YFF1001400), the National Natural Science Foundation of China (32360509), the Natural Science Foundation of Xinjiang Uygur Autonomous Region (2024D01A150), the Key Research and Development Program of Xinjiang Autonomous Region (2023B02014), the Corps of Agricultural Science and Technology Innovation Project Special (NCG202316), Key Research and Development Program of Xinjiang (2022B02052), the National Natural Science Foundation of China (32301888), the National Key Laboratory of Cotton Bio-breeding and Integrated Utilization (CBIU2024004), and the Natural Science Foundation of Henan (232300421253).
About Plant Phenomics
Science Partner Journal Plant Phenomics is an online-only Open Access journal published in affiliation with the State Key Laboratory of Crop Genetics & Germplasm Enhancement, Nanjing Agricultural University (NAU) and distributed by the American Association for the Advancement of Science (AAAS). Like all partners participating in the Science Partner Journal program, Plant Phenomics is editorially independent from the Science family of journals. Editorial decisions and scientific activities pursued by the journal’s Editorial Board are made independently, based on scientific merit and adhering to the highest standards for accurate and ethical promotion of science. These decisions and activities are in no way influenced by the financial support of NAU, NAU administration, or any other institutions and sponsors. The Editorial Board is solely responsible for all content published in the journal. To learn more about the Science Partner Journal program, visit the SPJ program homepage.