By reconstructing 366 sorghum plants from calibrated images, the team showed that automated measurements achieved near-human repeatability and enabled genome-wide association studies (GWAS) to uncover genetic markers linked to leaf orientation.
Global food security depends heavily on crops that can thrive under high planting densities and limited water supplies. In maize, breeding for improved canopy structure has already enhanced productivity by redistributing light more evenly across leaves, reducing water loss through transpiration, and improving radiation use efficiency. However, while traits such as leaf angle, internode length, and plant height have been extensively studied, phyllotaxy has remained difficult to evaluate at scale due to labor-intensive measurement methods. Traditional approaches involve manual protractors, top-down imaging, or slow digitization methods, all of which limit throughput. Based on these challenges, there has been an urgent need for new, high-throughput phenotyping methods that can quantify phyllotaxy accurately and enable genetic studies.
A study (DOI: 10.1016/j.plaphe.2025.100023) published in Plant Phenomics on 8 March 2025 by James C. Schnable’s team, University of Nebraska-Lincoln, highlights the potential of 3D phenotyping to accelerate breeding for improved canopy architecture, with implications for enhancing light capture, water use efficiency, and yield in sorghum and other staple crops.
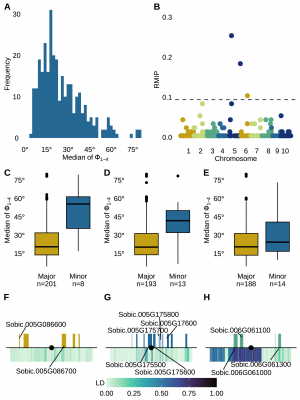
In this study, researchers employed an automated 3D reconstruction method to evaluate phyllotaxy—the angular arrangement of leaves—across diverse sorghum genotypes. Traditional manual measurement of each leaf angle requires 10–20 minutes per plant, limiting throughput. By contrast, the 3D voxel-based approach reconstructed plants from calibrated images and extracted angles with far less labor. To assess reliability, comparisons were made between manual and automated datasets. Manual measurements by two individuals achieved a correlation of R² = 0.55, while automated measurements from images collected two days apart reached R² = 0.41. When time gaps extended to three or five days, correlations dropped to R² = 0.33 and R² = 0.24, respectively, suggesting sensitivity to plant growth dynamics. Automated reconstructions also showed a moderate correlation (R² = 0.48) with the combined manual dataset, approaching human-level repeatability. Applying this method to 336 sorghum plants representing 236 genotypes revealed broad variation in phyllotaxy, with angles ranging from nearly perfect alternation (Φ₂ = 1.05°) to nearly overlapping leaves (Φ₂ = 170.4°). While some extreme values were validated through manual image inspection, many deviations beyond 90° proved unreliable due to issues such as tillers, senescing leaves, or leaf ordering errors, and these were excluded from analysis. Beyond phenotyping, the researchers conducted a genome-wide association study (GWAS) using 25 quantitative phyllotaxy metrics, 13 of which showed heritability estimates ≥0.2. Seven metrics yielded stable marker-trait associations, highlighting six genomic regions repeatedly linked to leaf arrangement. The most robust signal involved marker Chr05:12,109,370, associated with the median phyllotaxic angle of the lower canopy, and additional markers on chromosomes 5 and 6 implicated genes encoding transcription factors, kinases, and stress-response proteins. Together, these results demonstrate that automated 3D imaging offers a reliable and scalable tool for dissecting the genetic basis of phyllotaxy in sorghum, advancing both functional genomics and breeding for improved canopy architecture.
The ability to rapidly and accurately measure phyllotaxy creates opportunities to improve crop breeding in sorghum and potentially maize. Optimizing canopy architecture could lead to more efficient light capture, reduced water use, and higher yields, especially under climate stress conditions. By identifying genetic markers associated with favorable leaf arrangements, breeders may employ marker-assisted selection to introduce desirable traits more efficiently.
###
References
DOI
Original URL
https://doi.org/10.1016/j.plaphe.2025.100023
Funding information
This work was supported by the Foundation for Food and Agriculture Research (602757), USDA-NIFA (2020-68013-32371 and 2024-67013-42449), Department of Energy the Office of Science (BER), U.S. DOE (DE-SC0020355), the National Science Foundation (IOS-2412930, 2417510, and 2412928), and the University of Nebraska-Lincoln’s Complex Biosystems Graduate Program. JMD is supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. 2034837.
About Plant Phenomics
Science Partner Journal Plant Phenomics is an online-only Open Access journal published in affiliation with the State Key Laboratory of Crop Genetics & Germplasm Enhancement, Nanjing Agricultural University (NAU) and distributed by the American Association for the Advancement of Science (AAAS). Like all partners participating in the Science Partner Journal program, Plant Phenomics is editorially independent from the Science family of journals. Editorial decisions and scientific activities pursued by the journal’s Editorial Board are made independently, based on scientific merit and adhering to the highest standards for accurate and ethical promotion of science. These decisions and activities are in no way influenced by the financial support of NAU, NAU administration, or any other institutions and sponsors. The Editorial Board is solely responsible for all content published in the journal. To learn more about the Science Partner Journal program, visit the SPJ program homepage.