Lane N, Power. Sex, Suicide: Mitochondria and the Meaning of Life. 2nd edition. Oxford, UK: Oxford University Press; 2018.
Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–97. https://doi.org/10.1146/annurev.genet.33.1.351.
Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20:745–54. https://doi.org/10.1038/s41556-018-0124-1.
Stampar SN, Broe MB, Macrander J, Reitzel AM, Brugler MR, Daly M. Linear mitochondrial genome in anthozoa (Cnidaria): a case study in ceriantharia. Sci Rep. 2019;9:6094. https://doi.org/10.1038/s41598-019-42621-z.
Arafat H, Alamaru A, Gissi C, Huchon D. Extensive mitochondrial gene rearrangements in ctenophora: insights from benthic platyctenida. BMC Evol Biol. 2018;18:65. https://doi.org/10.1186/s12862-018-1186-1.
Cameron SL, Yoshizawa K, Mizukoshi A, Whiting MF, Johnson KP. Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera). BMC Genomics. 2011;12:394. https://doi.org/10.1186/1471-2164-12-394.
Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–80. https://doi.org/10.1093/nar/27.8.1767.
Boore JL, Macey JR, Medina M. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol. 2005;395:311–48. https://doi.org/10.1016/S0076-6879(05)95019-2.
Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–35. https://doi.org/10.1146/annurev.biochem.66.1.409.
Pereira SL. Mitochondrial genome organization and vertebrate phylogenetics. Genet Mol Biol. 2000;23:745–52. https://doi.org/10.1590/S1415-47572000000400008.
Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 2014;59:95–117. https://doi.org/10.1146/annurev-ento-011613-162007.
Marcus JM. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genet. 2018;05:001–23. https://doi.org/10.3934/genet.2018.1.1.
Shao R, Barker SC. Mitochondrial genomes of parasitic arthropods: implications for studies of population genetics and evolution. Parasitology. 2007;134:153–67. https://doi.org/10.1017/S0031182006001429.
Smith DR. The past, present and future of mitochondrial genomics: have we sequenced enough mtdnas? Brief Funct Genomics. 2015;elv027. https://doi.org/10.1093/bfgp/elv027.
Zardoya R. Recent advances in Understanding mitochondrial genome diversity. F1000Res. 2020;9:1–19. https://doi.org/10.12688/f1000research.21490.1.
Shields O. World distribution of the Vanessa Cardui group (Nymphalidae). J Lepid Soc. 1992;46:235–8.
Newland D, Still R, Swash A, Tomlinson D. Britain’s Butterflies: A Field Guide to the Butterflies of Great Britain and Ireland. 3rd edition. New Jersey: Princeton University Press; 2015.
Stefanescu C, Páramo F, Åkesson S, Alarcón M, Ávila A, Brereton T, et al. Multi-generational long-distance migration of insects: studying the painted lady butterfly in the Western palaearctic. Ecography. 2013;36:474–86. https://doi.org/10.1111/j.1600-0587.2012.07738.x.
Rajaei H, Aarvik L, Arnscheid WR, Baldizzone G, Bartsch D, Bengtsson BÅ, et al. Catalogue of the lepidoptera of Iran*. Integr Syst. 2023;6:121–459. https://doi.org/10.18476/2023.997558.7.
Johnson G, Butterfly expedition to, Iran. 1966. The entomologist’s record and journal of variation. 1967;79:35–42.
Reed RD, Nagy LM. Evolutionary redeployment of a biosynthetic module: expression of eye pigment genes vermilion, cinnabar, and white in butterfly wing development. Evol Dev. 2005;7:301–11. https://doi.org/10.1111/j.1525-142X.2005.05036.x.
Abbasi R, Marcus JM. Color pattern evolution in Vanessa butterflies (Nymphalidae: Nymphalini): non-eyespot characters. Evol Dev. 2015;17:63–81. https://doi.org/10.1111/ede.12109.
Abbasi R, Marcus JM. Colour pattern homology and evolution in Vanessa butterflies (Nymphalidae: Nymphalini): eyespot characters. J Evol Biol. 2015;28:2009–26. https://doi.org/10.1111/jeb.12716.
Abbasi R, Marcus JM. A new A-P compartment boundary and organizer in holometabolous insect wings. Sci Rep. 2017;7:16337. https://doi.org/10.1038/s41598-017-16553-5.
Naderi A. The field guide of butterflies of Iran. Tehran-Iran: Iranshenasi; 2019.
Liu G, Chang Z, Chen L, He J, Dong Z, Yang J, et al. Genome size variation in butterflies (Insecta, Lepidotera, Papilionoidea): a thorough phylogenetic comparison. Syst Entomol. 2020;45:571–82. https://doi.org/10.1111/syen.12417.
Park JS, Kim MJ, Jeong SY, Kim SS, Kim I. Complete mitochondrial genomes of two gelechioids, mesophleps albilinella and dichomeris Ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in lepidoptera. Curr Genet. 2016;62:809–26. https://doi.org/10.1007/s00294-016-0585-3.
Liao F, Wang L, Wu S, Li Y-P, Zhao L, Huang G-M, et al. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 2010;172–86. https://doi.org/10.7150/ijbs.6.172.
Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–5. https://doi.org/10.1093/bioinformatics/btm573.
Boore JL, Lavrov DV, Brown WM. Gene translocation links insects and crustaceans. Nature. 1998;392(6677):667–8. https://doi.org/10.1038/33577.
Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci U S A. 2012;109:19333–8. https://doi.org/10.1073/pnas.1213199109.
Tamura K, Tao Q, Kumar S. Theoretical foundation of the reltime method for estimating divergence times from variable evolutionary rates. Mol Biol Evol. 2018;35:1770–82. https://doi.org/10.1093/molbev/msy044.
Nei M, Kumar S. Molecular Evolution and Phylogenetics. 1st edition. New York: Oxford University Press; 2000.
Tao Q, Tamura K, Mello B, Kumar S. Reliable confidence intervals for reltime estimates of evolutionary divergence times. Mol Biol Evol. 2020;37:280–90. https://doi.org/10.1093/molbev/msz236.
Formenti G, Rhie A, Balacco J, Haase B, Mountcastle J, Fedrigo O, et al. Complete vertebrate mitogenomes reveal widespread repeats and gene duplications. Genome Biol. 2021;22:120. https://doi.org/10.1186/s13059-021-02336-9.
Borisova OF, Shchyolkina AK, Chernov BK, Tchurikov NA. Relative stability of AT and GC pairs in parallel DNA duplex formed by a natural sequence. FEBS Lett. 1993;322:304–6. https://doi.org/10.1016/0014-5793(93)81591-M.
Hu E-Z, Lan X-R, Liu Z-L, Gao J, Niu D-K. A positive correlation between GC content and growth temperature in prokaryotes. BMC Genomics. 2022;23:110. https://doi.org/10.1186/s12864-022-08353-7.
Bernardi G, Bernardi G. Compositional constraints and genome evolution. J Mol Evol. 1986;24:1–11. https://doi.org/10.1007/BF02099946.
Church BV, Williams HT, Mar JC. Investigating skewness to understand gene expression heterogeneity in large patient cohorts. BMC Bioinformatics. 2019;20(S24):668. https://doi.org/10.1186/s12859-019-3252-0.
Frank AC, Lobry JR. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene. 1999;238:65–77. https://doi.org/10.1016/S0378-1119(99)00297-8.
Hubert B. SkewDB, a comprehensive database of GC and 10 other skews for over 30,000 chromosomes and plasmids. Sci Data. 2022;9:92. https://doi.org/10.1038/s41597-022-01179-8.
Montaña-Lozano P, Moreno-Carmona M, Ochoa-Capera M, Medina NS, Boore JL, Prada CF. Comparative genomic analysis of vertebrate mitochondrial reveals a differential of rearrangements rate between taxonomic class. Sci Rep. 2022;12:5479. https://doi.org/10.1038/s41598-022-09512-2.
Mwinyi A, Meyer A, Bleidorn C, Lieb B, Bartolomaeus T, Podsiadlowski L. Mitochondrial genome sequence and gene order of sipunculus nudus give additional support for an inclusion of sipuncula into annelida. BMC Genomics. 2009;10:27. https://doi.org/10.1186/1471-2164-10-27.
Shtolz N, Mishmar D. The metazoan landscape of mitochondrial DNA gene order and content is shaped by selection and affects mitochondrial transcription. Commun Biol. 2023;6:93. https://doi.org/10.1038/s42003-023-04471-4.
Wahlberg N, Braby MF, Brower AVZ, de Jong R, Lee M-M, Nylin S, et al. Synergistic effects of combining morphological and molecular data in resolving the phylogeny of butterflies and skippers. Proc Biol Sci. 2005;272:1577–86. https://doi.org/10.1098/rspb.2005.3124.
Yan Z-T, Fan Z-H, He S-L, Wang X-Q, Chen B, Luo S-T. Mitogenomes of eight nymphalidae butterfly species and reconstructed phylogeny of nymphalidae (Nymphalidae: Lepidoptera). Genes. 2023;14:1018. https://doi.org/10.3390/genes14051018.
Freitas AVL, Brown KS. Phylogeny of the nymphalidae (Lepidoptera). Syst Biol. 2004;53(3):363–83. https://doi.org/10.1080/10635150490445670.
Lang S-Y. The nymphalidae of China (Lepidoptera, Rhopalocera). In: Libytheinae PI, editor. Danainae, Calinaginae, Morphinae, Heliconninae, Nymphalinae, Charaxinae, Apaturinae, Cyrestinae, Biblidinae, limenitinae. Pardubice, Czech Republic: Tshikolovets; 2012.
Zhang H, Chen Q, Xie Q, Lin Q, Sun G, Fang Y, et al. The complete mitochondrial genome of Stibochiona Nicea (Gray, 1846) (Lepidoptera: Nymphalidae) and phylogenetic analysis. Mitochondr DNA Part B. 2023;8:648–52. https://doi.org/10.1080/23802359.2023.2221348.
Brower AVZ. Phylogenetic relationships among the Nymphalidae (Lepidoptera) inferred from partial sequences of the wingless gene. Proc R Soc Lond B Biol Sci. 2000;267:1201–11. https://doi.org/10.1098/rspb.2000.1129.
Wahlberg N, Weingartner E, Nylin S. Towards a better understanding of the higher systematics of nymphalidae (Lepidoptera: Papilionoidea). Mol Phylogenet Evol. 2003;28:473–84. https://doi.org/10.1016/S1055-7903(03)00052-6.
Wahlberg N, Brower AVZ, Nylin S. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily nymphalinae (Lepidoptera: Nymphalidae). Biol J Linn Soc. 2005;86:227–51. https://doi.org/10.1111/j.1095-8312.2005.00531.x.
Su C, Shi Q, Sun X, Ma J, Li C, Hao J, et al. Dated phylogeny and dispersal history of the butterfly subfamily nymphalinae (Lepidoptera: Nymphalidae). Sci Rep. 2017;7:8799. https://doi.org/10.1038/s41598-017-08993-w.
Wan X, Kim MJ, Cho Y, Jun J, Jeong HC, Lee KY, et al. Sequence divergence and phylogenetic investigation of the Nymphalidae (Lepidoptera: Papilionoidea) occurring in South Korea. Int J Ind Entomol. 2013;26:95–112. https://doi.org/10.7852/ijie.2013.26.2.95.
Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AVL, et al. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc R Soc Lond B Biol Sci. 2009;276:4295–302. https://doi.org/10.1098/rspb.2009.1303.
Peters MJ, Marcus JM. Taxonomy as a hypothesis: testing the status of the Bermuda Buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Entomol. 2017;42:288–300. https://doi.org/10.1111/syen.12214.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. https://doi.org/10.1093/bioinformatics/bts199.
Lu Y, Liu N, Xu L, Fang J, Wang S. The complete mitochondrial genome of Vanessa indica and phylogenetic analyses of the family nymphalidae. Genes Genomics. 2018;40:1011–22. https://doi.org/10.1007/s13258-018-0709-x.
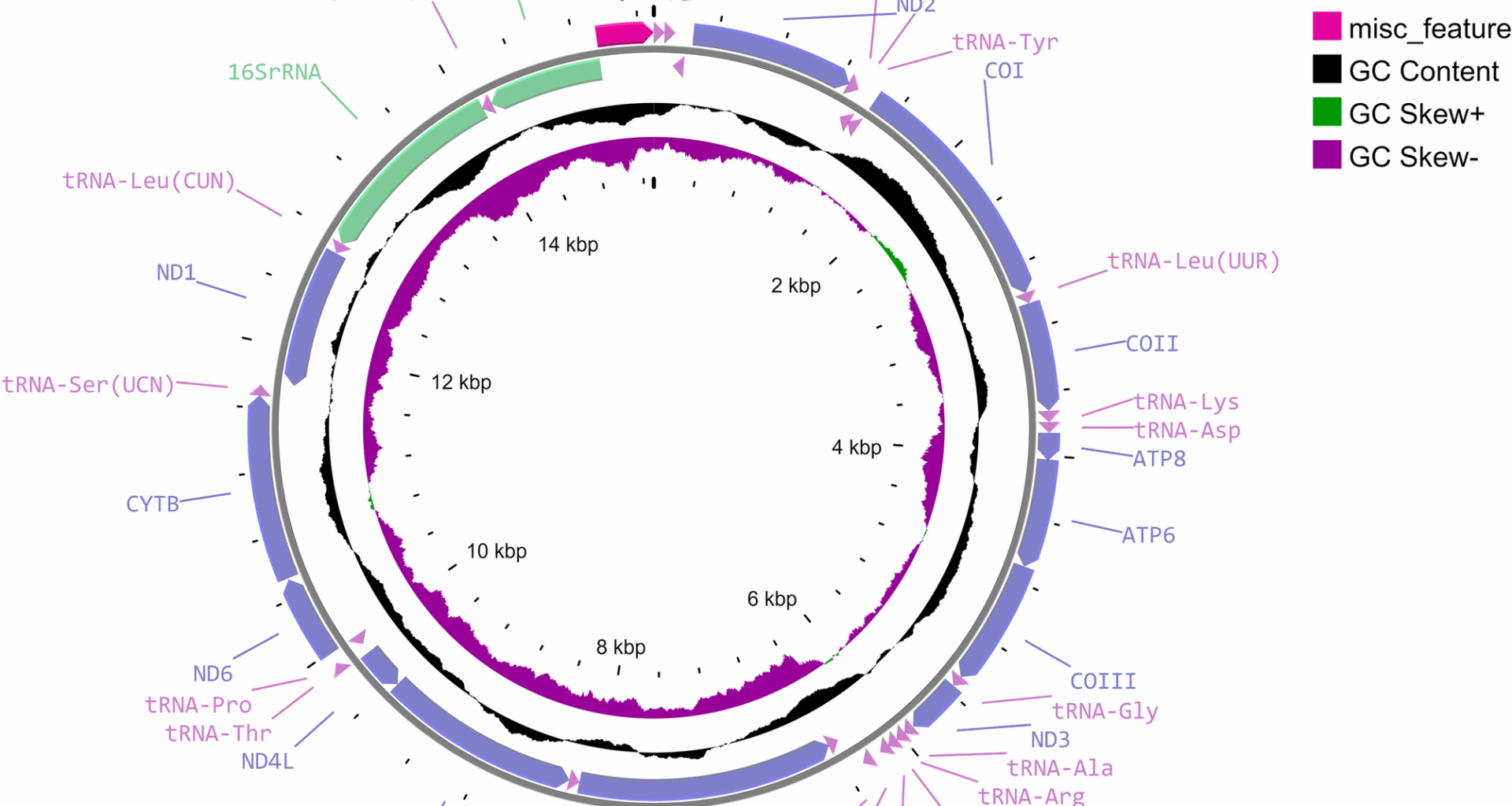
Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C, et al. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023;51:W484–92. https://doi.org/10.1093/nar/gkad326.
Perna NT, Kocher ThomasD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 1995;41. https://doi.org/10.1007/BF00186547.
Sharp PM, Tuohy TMF, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–43. https://doi.org/10.1093/nar/14.13.5125.
Khandia R, Singhal S, Kumar U, Ansari A, Tiwari R, Dhama K, et al. Analysis of Nipah virus codon usage and adaptation to hosts. Front Microbiol. 2019. https://doi.org/10.3389/fmicb.2019.00886.
Ackery PR, de Jong R, Vane-Wright RI. 16. The butterflies: Hedyloidea, hesperioidea and papilionoidea. Band 4: Arthropoda, 2 Hälfte: Insecta, Lepidoptera, moths and Butterflies, Teilband/Part 35, volume 1: Evolution, Systematics, and biogeography. DE GRUYTER; 1998. pp. 263–300. https://doi.org/10.1515/9783110804744.263.
Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–77. https://doi.org/10.1080/10635150701472164.
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52. https://doi.org/10.1093/oxfordjournals.molbev.a026334.
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36. https://doi.org/10.1093/nar/gkn180. Web Server:W465–9.
Alexiuk MR, Lalonde MML. Phylogenetic analysis of the complete mitochondrial genome of the blomfild’s beauty butterfly Smyrna Blomfildia (Fabricius 1781) (Insecta: lepidoptera: nymphalidae: Nymphalini). Mitochondr DNA Part B. 2021;6:3199–201. https://doi.org/10.1080/23802359.2021.1989337.
Agcaoili AM, Ameena N, Andres D, Caners R, Chahal MK, Croitor NJ, et al. Phylogenetic analysis of the complete mitochondrial genome of the orange-winged sulphur butterfly Dercas nina mell 1913 (Insecta: lepidoptera: pieridae: Coliadinae). Mitochondrial DNA Part B. 2024;9:1510–7. https://doi.org/10.1080/23802359.2024.2427109.
Alexiuk MR, Lalonde MML, Marcus JM. Phylogenetic analysis of the complete mitochondrial genome of the Japanese Peacock butterfly Aglais Io geisha (Stichel 1907) (Insecta: lepidoptera: Nymphalidae). Mitochondr DNA B Resour. 2021;6:3082–4. https://doi.org/10.1080/23802359.2021.1981168.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. https://doi.org/10.1093/nar/22.22.4673.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal w and clustal x version 2.0. Bioinformatics. 2007;23:2947–8. https://doi.org/10.1093/bioinformatics/btm404.
Kumar S, Stecher G, Suleski M, Sanderford M, Sharma S, Tamura K. MEGA12: molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol Biol Evol. 2024. https://doi.org/10.1093/molbev/msae263.
Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–23. https://doi.org/10.1109/TAC.1974.1100705.
Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–4.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. https://doi.org/10.1093/sysbio/sys029.
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 2018;67:901–4. https://doi.org/10.1093/sysbio/syy032.
Rambaut A. FigTree. 2023.
Hall TA, BioEdit:. A User-Friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. https://doi.org/10.14601/Phytopathol_Mediterr-14998u1.29.
Milec LJM, Vanhove MPM, Bukinga FM, De Keyzer ELR, Kapepula VL, Masilya PM, et al. Complete mitochondrial genomes and updated divergence time of the two freshwater clupeids endemic to lake Tanganyika (Africa) suggest intralacustrine speciation. BMC Ecol Evol. 2022;22:127. https://doi.org/10.1186/s12862-022-02085-8.
Mello B. Estimating timetrees with MEGA and the timetree resource. Mol Biol Evol. 2018;35:2334–42. https://doi.org/10.1093/molbev/msy133.
Kumar S, Stecher G, Suleski M, Hedges SB, TimeTree. A resource for Timelines, Timetrees, and divergence times. Mol Biol Evol. 2017;34:1812–9. https://doi.org/10.1093/molbev/msx116.