During 2021–2022, faecal samples were obtained from 144 lizards (agamas, geckos, iguanas, skinks), 131 snakes, 30 tortoises, and 3 crocodiles (for more details, see Additional file 1). The samples originated from 12 private breeding facilities, 4 zoological gardens, and 2 pet stores of 1 chain (for more details, see Additional file 2). Of the 308 examined individuals, 55 individuals (17 lizards, 38 snakes, 0 tortoises, and 0 crocodiles) were microscopically positive for coccidian parasites, and 10 individuals (9 lizards, 1 snake, 0 tortoises, and 0 crocodiles) for Cryptosporidium (Additional files 1, 3, 4). Four genera of coccidian parasites were recorded: Caryospora, Choleoeimeria, Eimeria, and Isospora. Photomicrographs of selected oocysts are presented in Additional file 5.
Coccidia
The final alignment of 18S rDNA yielded a 1,378 bp Long matrix, containing 112 coccidian sequences; 50 sequences originated from reptiles. The final alignment of COI was 693 bp Long and contained 57 coccidian sequences, 11 of which originated from reptiles.
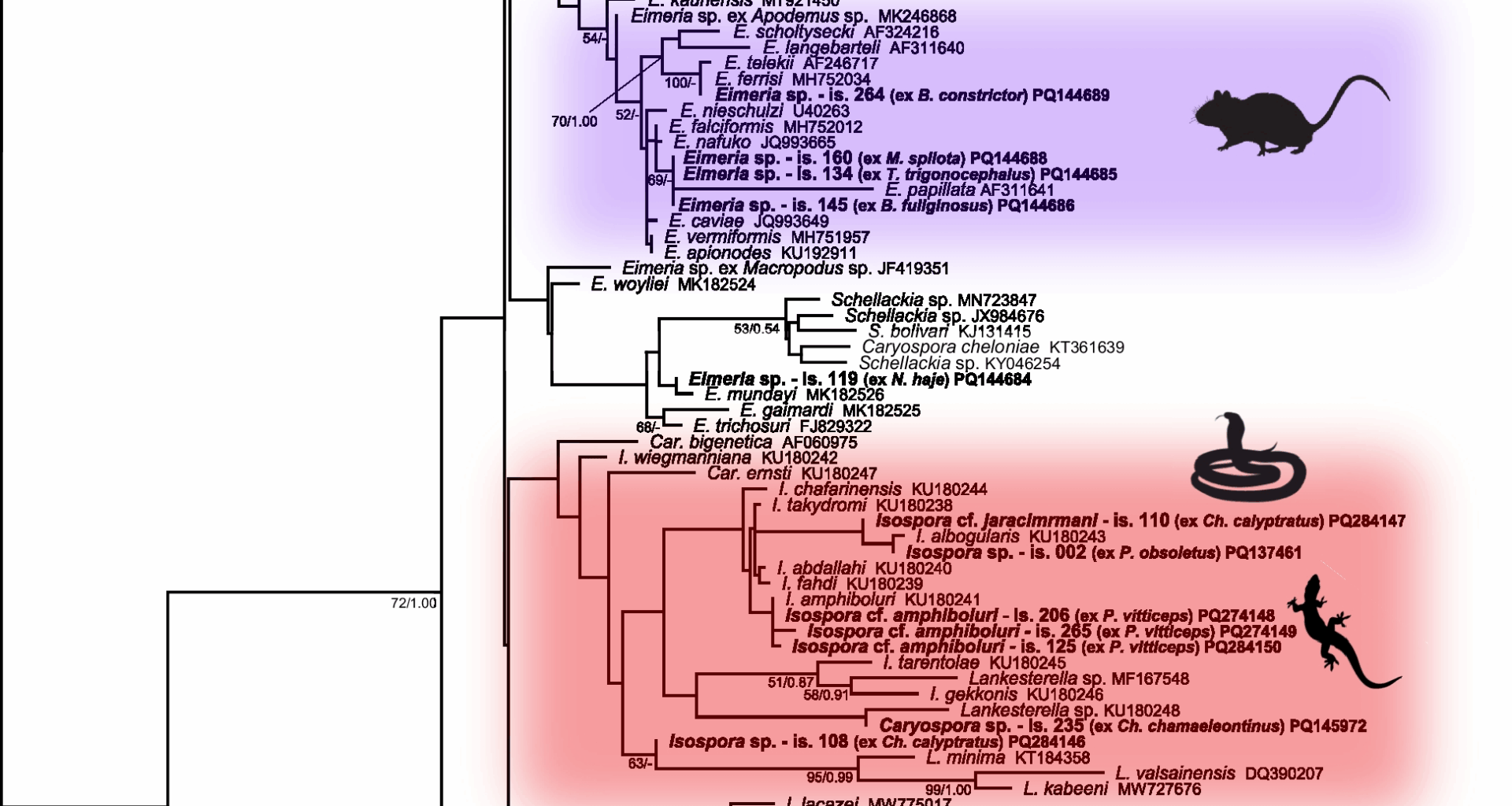
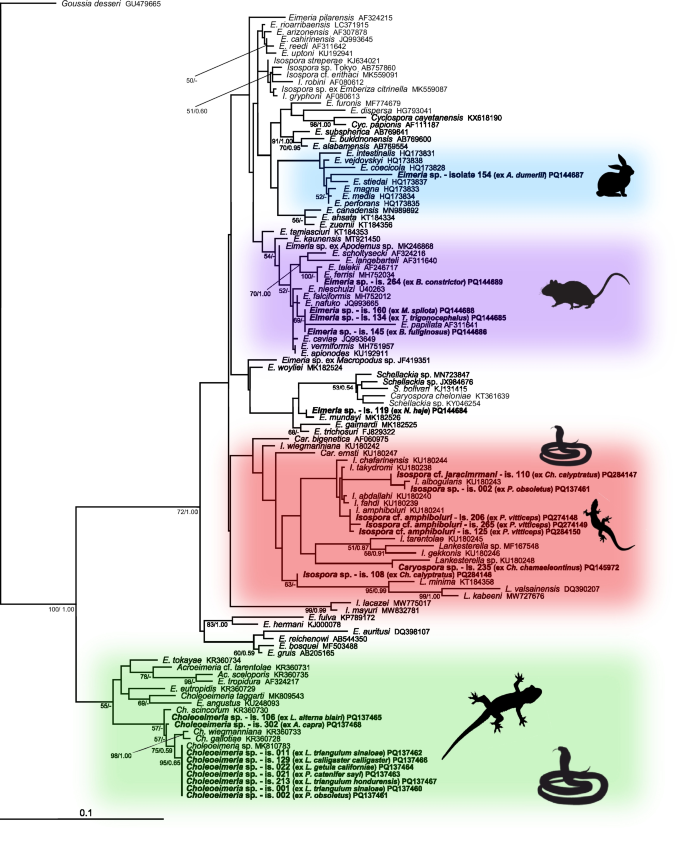
Compared to COI sequences, 18S rDNA was amplified more successfully (Additional file 6): nine sequences of Choleoeimeria were obtained from seven snake species and one agamid lizard (Fig. 1). Five sequences of Isospora were obtained from chameleons and agamas, morphologically corresponding to Isospora jaracimrmani and Isospora amphiboluri, respectively. A single Caryospora sequence was obtained from an iguanid lizard (Chamaeleolis chamaeleontinus). In addition, six sequences of the genus Eimeria were identified from booid and colubroid snakes; however, they all clustered within the rodent and rabbit eimerians, suggesting that these eimerians were only a passage from prey (Fig. 1).
Phylogenetic relationships inferred from the ML analysis of the 18S rDNA sequences of coccidia. The BI tree was mapped onto the ML tree. Numbers at the nodes show bootstrap values derived from the ML analysis and posterior probabilities from the BI analysis, shown as ML/BI. Only bootstrap supports and posterior probabilities higher than 50% or 0.50, respectively, are displayed. Bootstrap supports and posterior probabilities lower than 50% or 0.50, respectively, are marked with a dash (-). Taxa and their corresponding isolates are labeled as listed in the GenBank database. The tree is rooted with Goussia. The scale bar indicates the number of substitutions per site.
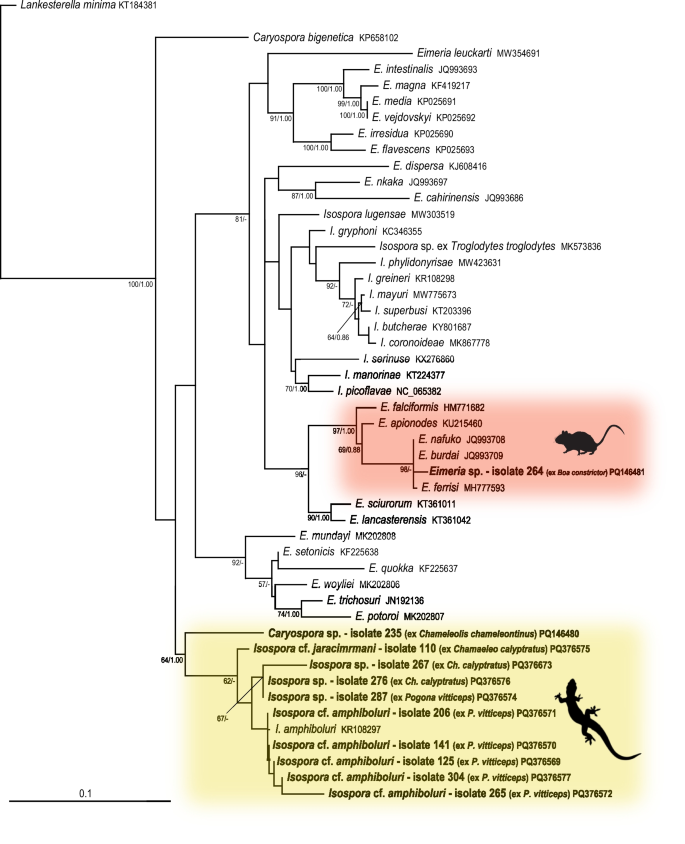
Eleven COI sequences were obtained from the examined samples (Additional file 6). Six sequences of Isospora were obtained from bearded dragons (Pogona vitticeps), five of them clustering with I. amphiboluri (KR108297) from the GenBank database. Three sequences were obtained from veiled chameleons (Chamaeleo calyptratus), one of them (sample number 110) morphologically corresponded to I. jaracimrmani. One Caryospora sequence was obtained from an iguanid lizard Ch. chamaeleontinus; the sequence originated from the same sample as the 18S rDNA sequence. One sequence of Eimeria was also obtained, originating from the same sample (number 264) as the 18S rDNA sequence and clustering with eimerians from rodents, which again indicated a passage from prey (Fig. 2).
Phylogenetic relationships inferred from the ML analysis of the COI sequences of coccidia. The BI tree was mapped onto the ML tree. Numbers at the nodes show bootstrap values derived from the ML analysis and posterior probabilities from the BI analysis, shown as ML/BI. Only bootstrap supports and posterior probabilities higher than 50% or 0.50, respectively, are displayed. Bootstrap supports and posterior probabilities lower than 50% or 0.50, respectively, are marked with a dash (-). Taxa and their corresponding isolates are labeled as listed in the GenBank database. The tree is rooted with Lankesterella. The scale bar indicates the number of substitutions per site.
All isosporans detected in samples from bearded dragons (P. vitticeps) – samples numbered 125, 141, 206, 265, 287, and 304 (Additional file 5) – were morphologically identical to I. amphiboluri described by [10] from P. vitticeps (Additional file 7). This was also congruent with the results of phylogenetic analyses (Figs. 1, 2).
In veiled chameleons (Ch. calyptratus), the spectrum of coccidia was not so uniform. The sample 276 (Additional file 5) also morphologically corresponded to I. amphiboluri. The sample 110 (Additional file 5) morphologically corresponded to I. jaracimrmani described by [11] from Ch. calyptratus. The sample 267 (Additional file 5) was also morphologically similar to I. jaracimrmani, however, the oocyst size was smaller than in the original description. The sample 108 (Additional file 5) most closely resembled Isospora wildi described by [12] from flapneck chameleon (Chamaeleo dilepis), however, the oocyst size was smaller compared to its original description. In addition, this sample was also similar to Isospora brygooi [13], described from the panther chameleon (Furcifer pardalis) (Additional file 7).
Only one sample (number 002) containing Isospora was obtained from snakes at all, namely from the western rat snake (Pantherophis obsoletus). However, this Isospora did not correspond to any previously described coccidia of the genus Isospora from snakes. Based on morphology, it most closely resembled Isospora cenchoae described by [14] from the Amazon Basin tree snake (Imantodes lentiferus), however, its oocysts were considerably smaller (Additional file 8). This sample was phylogenetically most closely related to Isospora albogularis (Fig. 1), however, both the oocysts and sporocysts were smaller than its original description. Similarly, it was placed on a common branch with the sample number 110 (Fig. 1), which morphologically corresponded to I. jaracimrmani, however, these two samples also differed in oocyst and sporocyst size.
The oocysts in the sample 302 (Additional file 5) from Acanthosaura capra (Squamata: Agamidae) most closely resembled Eimeria cameronensis, which was described from the great anglehead lizard (Gonocephalus grandis) by [15] and shares the same geographical range with A. capra. However, they were also similar to oocysts of the Choleoeimeria glawi [13], which are slightly larger (Additional file 9), and its sequence also clustered within Choleoeimeria (Fig. 1). In the phylogenetic tree, sample 302 was most closely related to Choleoeimeria gallotiae, Choleoeimeria scincorum, and Choleoeimeria wiegmanniana; however, all of these species have larger oocysts and differ in shape [16].
Coccidia included in the samples numbered 001 (Lampropeltis triangulum sinaloae), 002 (P. obsoletus), 011 (L. t. sinaloae), 021 (Pituophis catenifer sayi), 022 (L. getula californiae), 106 (L. alterna blairi), 129 (L. calligaster calligaster), and 213 (L. t. hondurensis) (Additional file 5) were morphologically most similar to Eimeria zamenis described from Coluber sp. by [17], and also recorded in Coluber constrictor flaviventris, L. calligaster calligaster, L. getula helbrooki, L. triangulum triangulum, Masticophis flagellum flagellum, and Masticophis flagellum testaceus [18]. However, oocysts of Eimeria detected in the above mentioned samples were also similar to several other Eimeria species, such as E. leptophis described from Leptophis mexicanus, E. oxybelis described from Oxybelis aenes, E. papillosum described from Salvadora grahaminae lineatus, E. scaphiodontophis described from Scaphiodontophis annulata, E. coniophanes described from Coniophanes fissidens, or to E. coniophis described from Conophis lineatus [19,20,21] (Additional file 10). In addition to the species mentioned above, the oocysts also resembled those of Choleoeimeria ghaffari described by [22] from Eryx jayakari, but they were slightly larger than the original description. Phylogenetically, all sequences of coccidia from the above mentioned samples clustered within Choleoeimeria spp. from the GenBank database (Fig. 1).
Sequences of coccidia included in samples 134 (Trimeresurus trigonocephalus), 145 (Boaedon fuliginosus), 154 (Acrantophis dumerili), 160 (Morelia spilota), and 264 (Boa constrictor) (Additional file 6) originated from unsporulated oocysts and clustered with Eimeria species described from rodents and rabbits (Fig. 1). Therefore, they most likely represented a passage from ingested (fed) rodents and rabbits. The sequence from the sample 154, obtained from A. dumerili, clustered together with sequences from domestic rabbits; the owner confirmed that the snake was frequently fed rabbits. Oocysts from the sample 264 (Additional file 5) were morphologically identical to Eimeria ferrisi described from the house mouse (Mus musculus) by [23]. Samples 134, 145, and 160 (Additional file 5) clustered with Eimeria papillata also described from M. musculus [24]. However, they were also morphologically similar in shape and size to Eimeria nieschulzi and E. vermiformis described from M. musculus by [24, 25]. All of the above mentioned snakes were routinely fed mice. As all these sequences originated from unsporulated oocysts, it was not possible to assign them morphologically to a particular species.
Regarding clinical manifestations observed in the examined animals, three unrelated individuals of P. vitticeps from different breeds exhibited reduced food intake, decreased activity, and suppressed growth compared to other animals within the same breeds. One of them was infected with I. amphiboluri, and two harboured a coinfection of I. amphiboluri with Ch. pogonae. Two individuals were treated with anticoccidials; their condition improved, and these animals survived. The third individual was not subjected to any therapy and died two months after the onset of clinical signs. Other individuals in this study that tested positive for coccidia showed no apparent clinical signs.
Cryptosporidium
Of the total number of examined faecal samples, oocysts of Cryptosporidium were microscopically detected in 10 samples (10/308; 3.2%) using the centrifugation-flotation concentration method (for more details, see Methods chapter and Additional file 1). DNA was isolated from all such microscopically positive samples, and amplified using three pairs of primers, each pair targeting a different gene. The final alignment of the actin gene was 822 bp Long and contained 29 sequences of Cryptosporidium; four of them originated from reptiles (Additional file 11). The final alignment of gp60 was 939 bp Long and contained 17 sequences of Cryptosporidium; one of them originated from reptiles (Additional file 11). Compared to other alignments analyzed in this study, the gp60 alignment contained so few sequences as only a limited number of sequences of this gene were available in the GenBank database. Unfortunately, amplification of the 18S rRNA gene was not successful.
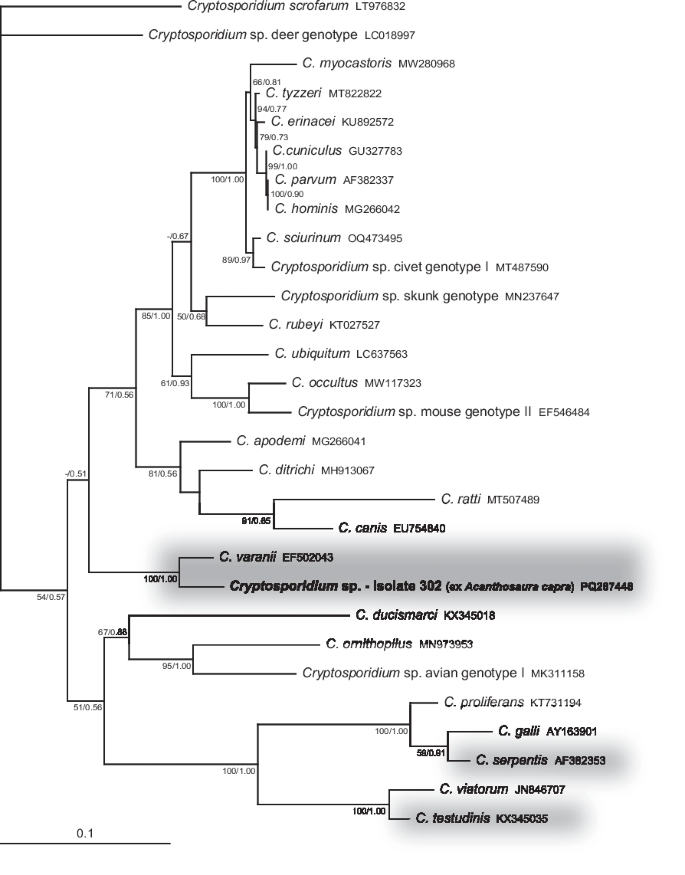
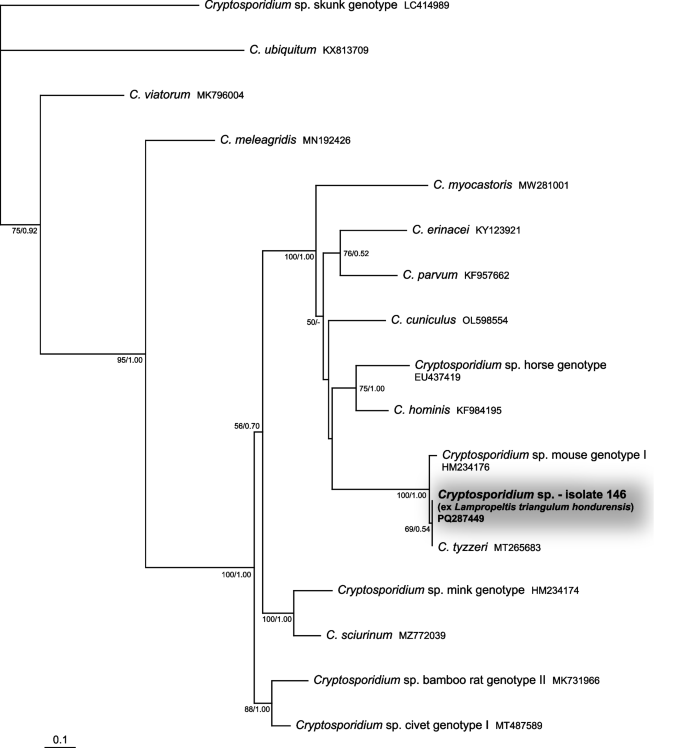
Only two Cryptosporidium sequences of sufficient quality were obtained. One was for the actin gene, originating from the green pricklenape A. capra (sample 302), which clustered together with the sequence of Cryptosporidium varanii from the GenBank database (EF502043) (Fig. 3). The morphology of the oocysts found in the faeces also corresponded to this species. The second sequence, targeting the gp60 gene, originated from the Honduran milksnake coral (L. triangulum hondurensis; sample 146). It clustered together with sequences from murine cryptosporidia (HM234176 and MT265683) (Fig. 4), which also corresponded with the oocyst size consistent with that of Cryptosporidium tyzzeri. Therefore, it again most likely represented a passage from fed rodents.
Phylogenetic relationships inferred from the ML analysis of the actin sequences of cryptosporidia. The BI tree was mapped onto the ML tree. Numbers at the nodes show bootstrap values derived from the ML analysis and posterior probabilities from the BI analysis, shown as ML/BI. Only bootstrap supports and posterior probabilities higher than 50% or 0.50, respectively, are displayed. Bootstrap supports and posterior probabilities lower than 50% or 0.50, respectively, are marked with a dash (-). Taxa and their corresponding isolates are labeled as listed in the GenBank database. The tree is rooted with Cryptosporidium scrofarum. The scale bar indicates the number of substitutions per site.
Phylogenetic relationships inferred from the ML analysis of the 60-kDa glycoprotein (gp60) sequences of cryptosporidia. The BI tree was mapped onto the ML tree. Numbers at the nodes show bootstrap values derived from the ML analysis and posterior probabilities from the BI analysis, shown as ML/BI. Only bootstrap supports and posterior probabilities higher than 50% or 0.50, respectively, are displayed. Bootstrap supports and posterior probabilities lower than 50% or 0.50, respectively, are marked with a dash (-). Taxa and their corresponding isolates are labeled as listed in the GenBank database. The tree is rooted with Cryptosporidium sp. skunk genotype. The scale bar indicates the number of substitutions per site.
A single individual of a leopard gecko (Eublepharis macularius), newly introduced to an established breeding group, presented with diarrhoea, apathy, and rapid weight loss despite continued food intake. A severe infection with Cryptosporidium was recorded, which was then introduced into the breeding group by this new animal due to the absence of proper quarantine. This individual, as well as four others from the infected breeding, was euthanised. No clinical symptoms were observed in the other individuals found to be infected with Cryptosporidium.