Zohary D, Hopf M. Domestication of plants in the old world: the origin and spread of cultivated plants in West asia, Europe and the nile Valley. 3rd ed. Oxford: Oxford University Press; 2000.
FAO. FAOSTAT: Crops and livestock products. Food and Agriculture Organization of the United Nations. 2023. Available from: https://www.fao.org/faostat [Accessed 22 Apr 2025].
Dhull SB, Kinabo J, Uebersax MA. Nutrient profile and effect of processing methods on the composition and functional properties of lentils (Lens culinaris Medik): a review. Legume Sci. 2023;5:e156. https://doi.org/10.1002/leg3.156.
Hossain A, Maitra S, Ahmed S, Mitra B, Ahmad Z, Garai S, Meena RS. Legumes for nutrient management in the cropping system. Advances in legumes for sustainable intensification. Amsterdam: Academic; 2022. pp. 93–112.
Shahzad A, Ullah S, Dar AA, Sardar MF, Mehmood T, Tufail MA, Haris M. Nexus on climate change: agriculture and possible solution to Cope future climate change stresses. Environ Sci Pollut Res. 2021;28:14211–32. https://doi.org/10.1007/s11356-020-11768-w.
Suri GK, Braich S, Noy DM, Rosewarne GM, Cogan NO, Kaur S. Advances in lentil production through heterosis: evaluating generations and breeding systems. PLoS ONE. 2022;17:e0262857. https://doi.org/10.1371/journal.pone.0262857.
Altaf MT, Cavagnaro PF, Kökten K, Ali A, Morales A, Tatar M, Baloch FS. Genotyping-by-sequencing derived SNP markers reveal genetic diversity and population structure of dactylis glomerata germplasm. Front Plant Sci. 2025;16:1530585.
Yalinkiliç NA, Başbağ S, Altaf MT, Ali A, Nadeem MA, Baloch FS. Applicability of scot markers in unraveling genetic variation and population structure among sugar beet (Beta vulgaris L.) germplasm. Mol Biol Rep. 2024;51:584.
Mohammed NA, Afzal M, Al-Faifi SA, Khan MA, Refay YA, Al-Samin BH, Ibrahim A. Effectiveness of sequence-related amplified polymorphism (SRAP) markers to assess the geographical origin and genetic diversity of collected lentil genotypes. Plant Biotechnol Rep. 2023;17:519–30. https://doi.org/10.1007/s11816-023-00792-6.
Marić S, Bolarić S, Martinčić J, Pejić I, Kozumplik V. Genetic diversity of hexaploid wheat cultivars estimated by RAPD markers, morphological traits and coefficients of parentage. Plant Breed. 2004;123:366–9. https://doi.org/10.1111/j.1439-0523.2004.00956.x.
Altaf MT, Tatar M, Ali A, Liaqat W, Mortazvi P, Kayihan C, Baloch FS. Advancements in QTL mapping and GWAS application in plant improvement. Turk J Bot. 2024;48:376–26.
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, et al. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip. 2018;32(2):261–85. https://doi.org/10.1080/13102818.2017.1400401.
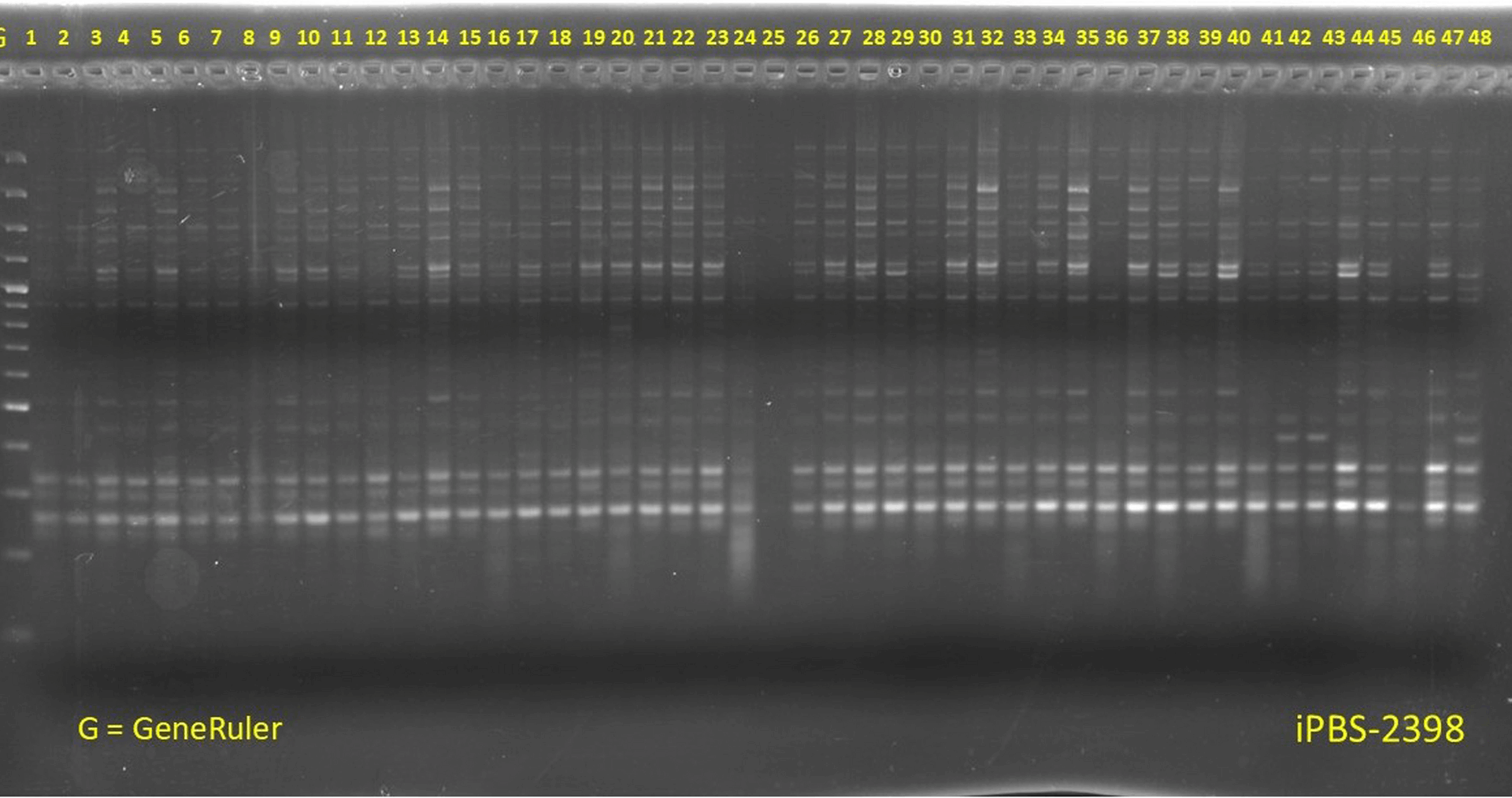
Kalendar R, Antonius K, Smýkal P, Schulman AH. iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor Appl Genet. 2010;121:1419–30. https://doi.org/10.1007/s00122-010-1398-2.
Uçer VA, Aglar E, Mortazavi P, Qureshi SA, Ali A, Tatar M, et al. Exploring genetic diversity of Turkish Fig (Ficus carica L.) germplasm using inter-primer binding site (iPBS) retrotransposon markers. Genet Resour Crop Evol. 2024. https://doi.org/10.1007/s10722-024-01654-2.
Madenova A, Kuan A, Ali A, Nadeem MA, Altaf MT, Kabylbekova B, Baloch FS. Exploring the population structure and genetic diversity in Apple germplasm using iPBS-retrotransposon markers. Turk J Bot. 2024;48:454–65. https://doi.org/10.55730/1300-008X.2767.
Baloch FS, Altaf MT, Bedir M, Nadeem MA, Tatar M, Karaköy T, Aasim M. iPBS-retrotransposons variations: DNA fingerprinting and the evaluation of genetic diversity and population structure in international Cowpea germplasm. Genet Resour Crop Evol. 2023;70:1867–77. https://doi.org/10.1007/s10722-023-01625-0.
Tahir NAR, Omer DA. (2017). Genetic variation in lentil genotypes by morphoagronomic traits and RAPD-PCR. The Journal of Animal and Plant Sciences. 2017;27(2):468-80.
Idrissi O, Udupa MS, De Keyser E, Van Damme P, De Riek J. Functional genetic diversity analysis and identification of associated simple sequence repeats and amplified fragment length polymorphism markers to drought tolerance in lentil (Lens Culinaris ssp. Culinaris Medicus) landraces. Plant Mol Biol Rep. 2016;34:659–80. https://doi.org/10.1007/s11105-015-0952-4.
Tomar S, Sharma S, Tripathi N, Thakur S, Pathak N, Sharma R, Tiwari P. SSR marker-based molecular characterization of lentil (Lens culinaris Medik.) genotypes. Legume Res. 2023;46:837–42. https://doi.org/10.18805/LR-6511.
Mammadova SE, Aghayeva SA. Genetic polymorphism assessment in a new lentil (Lens culinaris medik., 1787) collection using ISSR markers. Acta Biol Sib. 2023;9:1181–8. https://doi.org/10.14258/abs.v9.e65.
Yusuf M. Molecular characterization of some lentil cultivars using scot markers. Sci. 2017;7:840–7.
Baloch FS, Derya M, Andeden EE, Alsaleh A, Cömertpay G, Kilian B, Özkan H. Inter-primer binding site retrotransposon and inter-simple sequence repeat diversity among wild lens species. Biochem Syst Ecol. 2015;58:162–8. https://doi.org/10.1016/j.bse.2014.11.005.
Qureshi SA, Nadeem MA, Sarıkaya MF, Ali A, Tatar M, Altafl MT, et al. Unveiling genetic diversity and population structure in lentil (Lens culinaris) germplasm through scot markers. Mol Biol Rep. 2025;52(1):1–9. https://doi.org/10.1007/s11033-025-10876-7.
Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:39–40.
Schenk JJ, Becklund LE, Carey SJ, Fabre PP. What is the modified CTAB protocol? Characterizing modifications to the CTAB DNA extraction protocol. Appl Plant Sci. 2023;11:e11517. https://doi.org/10.1002/aps3.11517.
Yeh FC, Yang RC, Boyle TB, Ye ZH, Mao JX. POPGENE, the user-friendly shareware for population genetic analysis. Edmonton, AB, Canada: Molecular Biology and Biotechnology Centre, University of Alberta; 1997.
Peakall R, Smouse PE. GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–95. https://doi.org/10.1111/j.1471-8286.2005.01155.x.
R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. 2019. Available from: https://cir.nii.ac.jp/crid/1370298755636824325
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59.
Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20. https://doi.org/10.1111/j.1365-294X.2005.02553.x.
Altaf MT, Nadeem MA, Ali A, Liaqat W, Bedir M, Baran N, et al. Applicability of start codon targeted (SCoT) markers for the assessment of genetic diversity in bread wheat germplasm. Genet Resour Crop Evol. 2025;72:1205–18.
Rey-Baños R, Sáenz de Miera LE, García P, de la Pérez M. Obtaining retrotransposon sequences, analysis of their genomic distribution and use of retrotransposon-derived genetic markers in lentil (Lens culinaris Medik). PLoS ONE. 2017;12(4):e0176728. https://doi.org/10.1371/journal.pone.0176728.
Rajpal VR, Singh A, Kathpalia R, Thakur RK, Khan MK, Pandey A, et al. The prospects of gene introgression from crop wild relatives into cultivated lentil for climate change mitigation. Front Plant Sci. 2023;14:1127239. https://doi.org/10.3389/fpls.2023.1127239.
Kalbiyeva YE, Aghayeva SA, Duran ST, İsmayilova VM, Mammadov AM, Hasanova MY. Study of genetic polymorphism using ISSR markers in the lentil collection. J Surv Fish Sci. 2023;10:1918–23.
Sharma R, Chaudhary L, Kumar M. Microsatellites based assessment of genetic diversity and population structure of Indian lentil (Lens culinaris Medik.) genotypes. Biologia. 2023;78:2317–26. https://doi.org/10.1007/s11756-023-01355-6.
Sadık G, Yıldız M, Taşkın B, et al. Application of iPBS-retrotransposons markers for the assessment of genetic diversity and population structure among sugar beet (Beta vulgaris) germplasm from different regions of the world. Genet Resour Crop Evol. 2025;72(1):3039–49. https://doi.org/10.1007/s10722-024-02148-3.
Al-Mousa R, Abbas S, Alshaal A, Kountar K. ISSR analysis to detect genetic variation among some lentil genotypes in Syria. Al-Mukhtar J Sci. 2023;38(2):150–9. https://doi.org/10.54172/mjsc.v38i2.1204.
Baran N, Shimira F, Nadeem MA, et al. Exploring the genetic diversity and population structure of upland cotton germplasm by iPBS-retrotransposons markers. Mol Biol Rep. 2023;50:4799–811. https://doi.org/10.1007/s11033-023-08399-0.
Palaz EB, Demirel F, Adali S, et al. Genetic relationships of salep Orchid species and gene flow among Serapias vomeracea × Anacamptis morio hybrids. Plant Biotechnol Rep. 2023;17:315–27. https://doi.org/10.1007/s11816-022-00782-w.
Haliloğlu K, Türkoğlu A, Öztürk A, Niedbała G, Niazian M, Wojciechowski T, Piekutowska M. Genetic diversity and population structure in bread wheat germplasm from Türkiye using iPBS-retrotransposons-based markers. Agronomy. 2023;13:255. https://doi.org/10.3390/agronomy13010255.
Khazaei H, Caron CT, Fedoruk M, Diapari M, Vandenberg A, Coyne CJ, et al. Genetic diversity of cultivated lentil (Lens culinaris Medik.) and its relation to the world’s agro-ecological zones. Front Plant Sci. 2016;7:1093. https://doi.org/10.3389/fpls.2016.01093.
Nleya T, Vandenberg A, Walley FL, Deneke D. Lentil: Agronomy. In: Wrigley C, editor. Encyclopedia of Food Grains. 2nd ed. Amsterdam, The Netherlands: Academic Press; 2016. 223–30. ISBN: 978-0-12-394786-4. Publisher: Academic Press
Dhuppar P, Biyan S, Chintapalli B, Rao S. Lentil crop production in the context of climate change: an appraisal. Indian Res J Ext Educ. 2012;2(Special Issue):33–5.
Zohary D, Hopf M, Weiss E. Domestication of plants in the old world: the origin and spread of domesticated plants in Southwest asia, europe, and the mediterranean basin. 4th ed. Oxford, UK: Oxford University Press; 2012.
Coyne C, McGee R. Lentil. In: Singh M, editor. Genetic and genomic resources of grain legume improvement. Amsterdam, The Netherlands: Elsevier; 2013. pp. 157–80.
Davies PA, Lülsdorf MM, Ahmad M. Wild relatives and biotechnological approaches. In: Yadav SS, McNeil D, Stevenson PC, editors. Lentil: an ancient crop for modern times. Dordrecht, The Netherlands: Springer; 2007. pp. 225–40.
Liber M, Duarte I, Maia AT, Oliveira HR. The history of lentil (Lens culinaris subsp. culinaris) domestication and spread as revealed by genotyping-by-sequencing of wild and landrace accessions. Front Plant Sci. 2021;12:628439. https://doi.org/10.3389/fpls.2021.628439.
Šućur R, Ali A, Mortazavi P, Altaf MT, Tatar M, NadeemMA, Jocković B, Mladenov V, Fragkostefanakis S, Chung YS, Baloch FS. Exploring the genetic diversity and population structure of Serbian and selected European bread wheat cultivars through iPBS-retrotransposon markers Turk. J Agric for. 2025: 1–26.ŠUĆUR R, ALI A, MORTAZAVI P, ALTAF MT, TATAR M, NADEEM MA, JOCKOVIĆ B, MLADENOV V, FRAGKOSTEFANAKIS S, CHUNG YS, BALOCH FS. Exploring the genetic diversity and population structure of Serbian and selected European bread wheat cultivars through iPBS-retrotransposon markers. Turkish Journal of Agriculture and Forestry. 2025;49(4):661-72.
Altaf MT, Liaqat W, Ali A, Jamil A, Fahad M, Rahman MAU, et al. Advancing chickpea breeding: omics insights for targeted abiotic stress mitigation and genetic enhancement. Bioch Genet. 2025;63:1063–115.
Nadeem MA. Deciphering the genetic diversity and population structure of Turkish bread wheat germplasm using iPBS-retrotransposons markers. Mol Biol Rep. 2021;48:6739–48. https://doi.org/10.1007/s11033-021-06575-7.