Song S, Liu Y, Wang NR, Haney CH. Mechanisms in plant-microbiome interactions: lessons from model systems. Curr Opin Plant Biol. 2021 [cited 2022 Jun 5];62. Available from: https://pubmed.ncbi.nlm.nih.gov/33545444/.
Fitzpatrick CR, Salas-González I, Conway JM, Finkel OM, Gilbert S, Russ D, et al. The plant microbiome: from ecology to reductionism and beyond. Annu Rev Microbiol. 2020;74(1):81–100.
Xu L, Pierroz G, Wipf HML, Gao C, Taylor JW, Lemaux PG, et al. Holo-omics for deciphering plant-microbiome interactions. Microbiome. 2021 [cited 2022 Jun 5];9. Available from: https://pubmed.ncbi.nlm.nih.gov/33762001/.
Choi K, Khan R, Lee SW. Dissection of plant microbiota and plant-microbiome interactions. J Microbiol. 2021 [cited 2022 Jun 5];59:281–91. Available from: https://pubmed.ncbi.nlm.nih.gov/33624265/.
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–21. Available from: https://doi.org/10.1038/s41579-020-0412-1.
Vorholt JA, Vogel C, Carlström CI, Müller DB. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe. 2017;22:142–55.
Liu YX, Qin Y, Chen T, Lu M, Qian X, Guo X, et al. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell. 2021 [cited 2022 May 25];12:315–30. Available from: https://pubmed.ncbi.nlm.nih.gov/32394199/.
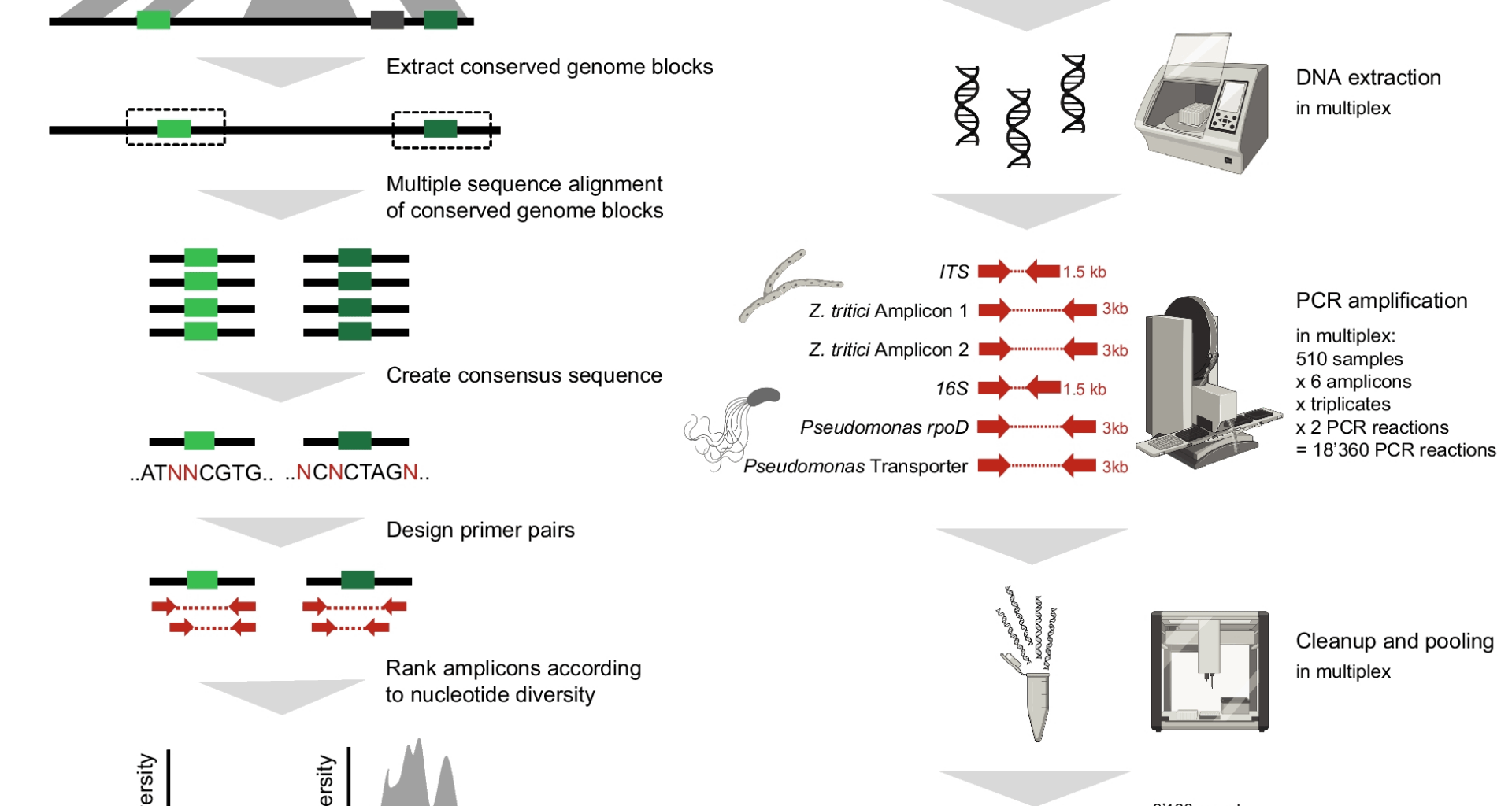
Hrovat K, Dutilh BE, Medema MH, Melkonian C. Taxonomic resolution of different 16S rRNA variable regions varies strongly across plant-associated bacteria. ISME Communications. 2024;4(1):ycae034
Earl JP, Adappa ND, Krol J, Bhat AS, Balashov S, Ehrlich RL, et al. Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific Biosciences sequencing of full-length 16S rRNA genes. Microbiome. 2018 [cited 2022 May 24];6. Available from: https://www.pmc/articles/PMC6199724/.
Wagner J, Coupland P, Browne HP, Lawley TD, Francis SC, Parkhill J. Evaluation of PacBio sequencing for full-length bacterial 16S rRNA gene classification. BMC Microbiol. 2016 [cited 2022 May 25];16:1–17. Available from: https://pubmed.ncbi.nlm.nih.gov/27842515/.
Callahan BJ, Wong J, Heiner C, Oh S, Theriot CM, Gulati AS, et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res . 2019 [cited 2022 May 25];47:E103. Available from: https://pubmed.ncbi.nlm.nih.gov/31269198/.
Escapa IF, Huang Y, Chen T, Lin M, Kokaras A, Dewhirst FE, et al. Construction of habitat-specific training sets to achieve species-level assignment in 16S rRNA gene datasets. Microbiome. 2020 [cited 2022 May 25];8:65. Available from: https://pubmed.ncbi.nlm.nih.gov/32414415/.
Cuscó A, Catozzi C, Viñes J, Sanchez A, Francino O. Microbiota profiling with long amplicons using nanopore sequencing: Full-length 16s rRNA gene and whole rrn operon . F1000Res. 2018;7:1755
Kinoshita Y, Niwa H, Uchida-Fujii E, Nukada T. Establishment and assessment of an amplicon sequencing method targeting the 16S-ITS-23S rRNA operon for analysis of the equine gut microbiome. Sci Rep. 2021;11:11884.
Wenger AM, Peluso P, Rowell WJ, Chang PC, Hall RJ, Concepcion GT, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nature Biotechnology 2019 37:10. 2019 [cited 2022 May 25];37:1155–62. Available from: https://www.nature.com/articles/s41587-019-0217-9.
Xin XF, Kvitko B, He SY. Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol. 2018;16:316–28.
Großkinsky DK, Tafner R, Moreno M V., Stenglein SA, De Salamone IEG, Nelson LM, et al. Cytokinin production by Pseudomonas fluorescens G20–18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis. Sci Rep. 2016 [cited 2022 May 25];6. Available from: https://pubmed.ncbi.nlm.nih.gov/26984671/.
Gislason AS, de Kievit TR. Friend or foe? Exploring the fine line between Pseudomonas brassicacearum and phytopathogens. J Med Microbiol. 2020 [cited 2024 Feb 20];69:347–60. Available from: https://pubmed.ncbi.nlm.nih.gov/31976855/.
Zboralski A, Filion M. Genetic factors involved in rhizosphere colonization by phytobeneficial Pseudomonas spp. Comput Struct Biotechnol J. 2020 [cited 2024 Feb 20];18:3539. Available from: https://www.pmc/articles/PMC7711191/.
Costa-Gutierrez SB, Adler C, Espinosa-Urgel M, de Cristóbal RE. Pseudomonas putida and its close relatives: mixing and mastering the perfect tune for plants. Appl Microbiol Biotechnol . 2022 [cited 2024 Feb 20];106:3351–67. Available from: https://pubmed.ncbi.nlm.nih.gov/35488932/.
Raio A, Puopolo G. Pseudomonas chlororaphis metabolites as biocontrol promoters of plant health and improved crop yield. World J Microbiol Biotechnol. 2021 [cited 2024 Feb 20];37. Available from: https://pubmed.ncbi.nlm.nih.gov/33978868/.
Weller DM. Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology. 2007 [cited 2024 Feb 20];97:250–6. Available from: https://pubmed.ncbi.nlm.nih.gov/18944383/.
Passera A, Compant S, Casati P, Maturo MG, Battelli G, Quaglino F, et al. Not just a pathogen? Description of a Plant-Beneficial Pseudomonas syringae Strain. Front Microbiol. 2019;10:1409.
Bailly A, Weisskopf L. Mining the volatilomes of plant-associated microbiota for new biocontrol solutions. Front Microbiol. 2017 [cited 2024 Feb 20];8. Available from: https://pubmed.ncbi.nlm.nih.gov/28890716/.
Biessy A, Filion M. Phenazines in plant-beneficial Pseudomonas spp.: biosynthesis, regulation, function and genomics. Environ Microbiol. 2018 [cited 2024 Feb 20];20:3905–17. Available from: https://pubmed.ncbi.nlm.nih.gov/30159978/.
Kavamura VN, Mendes R, Bargaz A, Mauchline TH. Defining the wheat microbiome: towards microbiome-facilitated crop production. Comput Struct Biotechnol J. 2021 [cited 2022 May 24];19:1200. Available from: https://www.pmc/articles/PMC7902804/.
Barroso-Bergadà D, Massot M, Vignolles N, d’Arcier JF, Chancerel E, Guichoux E, Walker AS, Vacher C, Bohan DA, Laval V, Suffert F. Metagenomic Next-Generation Sequencing (mNGS) Data Reveal the Phyllosphere Microbiome of Wheat Plants Infected by the Fungal Pathogen Zymoseptoria tritici. Phytobiomes J. 2023;7(2):281–7.
Kerdraon L, Barret M, Laval V, Suffert F. Differential dynamics of microbial community networks help identify microorganisms interacting with residue-borne pathogens: The case of Zymoseptoria tritici in wheat. Microbiome. 2019;7(1):125.
Sapkota R, Jørgensen LN, Nicolaisen M. Spatiotemporal variation and networks in the mycobiome of the wheat canopy. Front Plant Sci. 2017;8:1357.
Chen J, Sharifi R, Khan MSS, Islam F, Bhat JA, Kui L, et al. Wheat microbiome: structure, dynamics, and role in improving performance under stress environments. Front Microbiol . 2021 [cited 2022 May 24];12. Available from: https://www.pmc/articles/PMC8793483/.
Seybold H, Demetrowitsch TJ, Hassani MA, Szymczak S, Reim E, Haueisen J, et al. A fungal pathogen induces systemic susceptibility and systemic shifts in wheat metabolome and microbiome composition. Nature Communications 2020 11:1. 2020 [cited 2022 May 24];11:1–12. Available from: https://www.nature.com/articles/s41467-020-15633-x.
Levy E, Eyal Z, Chet I. Suppression of septoria tritici blotch and leaf rust on wheat seedling leaves by pseudomonads. Plant Pathol. 1988 [cited 2022 May 24];37:551–7. Available from: https://onlinelibrary.wiley.com/doi/full/. https://doi.org/10.1111/j.1365-3059.1988.tb02114.x.
Large EC. Growth stages in cereals—illustration of the Feekes scale. Plant Pathol. 1954 [cited 2024 Feb 23];3:128–9. Available from: https://www.onlinelibrary.wiley.com/doi/full/. https://doi.org/10.1111/j.1365-3059.1954.tb00716.x.
Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, et al. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics. 2010;11:1–14.
Peix A, Ramírez-Bahena MH, Velázquez E. The current status on the taxonomy of Pseudomonas revisited: An update. Infection, Genetics and Evolution. 2018;57:106–16. Available from: https://doi.org/10.1016/j.meegid.2017.10.026.
Gomila M, Peña A, Mulet M, Lalucat J, García-Valdés E. Phylogenomics and systematics in Pseudomonas. Front Microbiol. 2015;6:1–13.
Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1):238.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004 [cited 2023 Jul 14];32:1792. Available from: https://www.pmc/articles/PMC390337/.
Rice P, Longden L, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000 [cited 2023 Jul 14];16:276–7. Available from: https://pubmed.ncbi.nlm.nih.gov/10827456/.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012 [cited 2023 Jul 14];40:e115. Available from: https://www.pmc/articles/PMC3424584/.
Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016 [cited 2023 Jul 14];2:e000056. Available from: https://www.microbiologyresearch.org/content/journal/mgen/. https://doi.org/10.1099/mgen.0.000056.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011 [cited 2022 Jun 2];27:2156–8. Available from: https://pubmed.ncbi.nlm.nih.gov/21653522/.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013 [cited 2023 Jul 14];30:772–80. Available from: https://doi.org/10.1093/molbev/mst010.
Katoh K, Misawa K, Kuma KI, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002 [cited 2023 Jul 14];30:3059–66. Available from: https://doi.org/10.1093/nar/gkf436.
Feurtey A, Lorrain C, McDonald MC, Milgate A, Solomon PS, Warren R, et al. A thousand-genome panel retraces the global spread and adaptation of a major fungal crop pathogen. Nat Commun . 2023 [cited 2023 Jul 14];14. Available from: https://pubmed.ncbi.nlm.nih.gov/36828814/.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
R Core Team. R: a language and environment for statistical computing. 2022 [cited 2023 Jul 13]. Available from: https://www.r-project.org/.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011 [cited 2022 May 30];17:10–2. Available from: https://journal.embnet.org/index.php/embnetjournal/article/view/200/479.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 2016 13:7. 2016 [cited 2022 May 30];13:581–3. Available from: https://www.nature.com/articles/nmeth.3869.
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014 [cited 2022 Jun 2];42:D643–8. Available from: https://academic.oup.com/nar/article/42/D1/D643/1061236.
Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019 [cited 2022 Jun 2];47:D259–64. Available from: https://academic.oup.com/nar/article/47/D1/D259/5146189.
Shen W, Le S, Li Y, Hu F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 2016 [cited 2022 Jun 2];11:e0163962. Available from: https://journals.plos.org/plosone/article?id=. https://doi.org/10.1371/journal.pone.0163962.
Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016 [cited 2022 Jun 2];44:D646–53. Available from: https://pubmed.ncbi.nlm.nih.gov/26578582/.
Singh NK, Karisto P, Croll D. Population-level deep sequencing reveals the interplay of clonal and sexual reproduction in the fungal wheat pathogen Zymoseptoria tritici. Microb Genom. 2021 [cited 2022 May 25];7:678. Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC8627204/.
Badet T, Oggenfuss U, Abraham L, McDonald BA, Croll D. A 19-isolate reference-quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici. BMC Biol. 2020;18:1–18.
Mirarab S, Nguyen N, Guo S, Wang LS, Kim J, Warnow T. PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J Comput Biol . 2015 [cited 2022 Jun 2];22:377. Available from: https://www.pmc/articles/PMC4424971/.
Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009 [cited 2022 Jun 2];26:1641. Available from: https://www.pmc/articles/PMC2693737/.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics . 2014 [cited 2023 Jul 13];30:1312–3. Available from: https://doi.org/10.1093/bioinformatics/btu033.
Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol . 2006 [cited 2022 Jun 2];23:254–67. Available from: https://pubmed.ncbi.nlm.nih.gov/16221896/.
Shimoyama Y. ANIclustermap: A tool for drawing ANI clustermap between all-vs-all microbial genomes. 2022. Available from: https://github.com/moshi4/ANIclustermap/.
Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–2.
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74.
Smith MR. Information theoretic generalized Robinson-Foulds metrics for comparing phylogenetic trees. Bioinformatics. 2020;36(20):5007–13.
Best DJ, Roberts DE. Algorithm AS 89: the upper tail probabilities of Spearman’s Rho. Appl Stat. 1975;24:377.
Oksanen Jari. vegan: Community Ecology Package. 2022. Available from: https://vegandevs.github.io/vegan/.
Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019 [cited 2023 Jul 13];35:526–8. Available from: https://doi.org/10.1093/bioinformatics/bty633.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013 [cited 2023 Jul 13];8:e61217. Available from: https://journals.plos.org/plosone/article?id=. https://doi.org/10.1371/journal.pone.0061217.
Kolde R. pheatmap: Pretty Heatmaps. 2019. Available from: https://cran.r-project.org/web/packages/pheatmap/.
Wei T, Simko V. R package ‘corrplot’: Visualization of a correlation matrix. 2021. Available from: https://cran.r-project.org/web/packages/corrplot/.
Curry E, Zhang H. Data integration, manipulation and visualization of phylogenetic trees . Data integration, manipulation and visualization of phylogenetic trees. Chapman and Hall/CRC; 2022 [cited 2023 Jul 13]. Available from: https://www.taylorfrancis.com/books/mono/. https://doi.org/10.1201/9781003279242/data-integration-manipulation-visualization-phylogenetic-trees-guangchuang-yu.
Yu G. Using ggtree to Visualize Data on Tree-Like Structures. Curr Protoc Bioinformatics. 2020 [cited 2023 Jul 13];69. Available from: https://pubmed.ncbi.nlm.nih.gov/32162851/.
Wickham H. ggplot2: Elegant graphics for data analysis. Cham: Springer International Publishing; 2016 [cited 2023 Jul 13]. Available from: http://link.springer.com/. https://doi.org/10.1007/978-3-319-24277-4.
Lalucat J, Mulet M, Gomila M, García-Valdés E. Genomics in bacterial taxonomy: Impact on the genus pseudomonas. Genes (Basel). 2020;11(2):139.
Flury P, Aellen N, Ruffner B, Péchy-Tarr M, Fataar S, Metla Z, et al. Insect pathogenicity in plant-beneficial pseudomonads: Phylogenetic distribution and comparative genomics. ISME J. 2016;10:2527–42.
Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nature Communications 2019 10:1. 2019 [cited 2023 Jul 15];10:1–11. Available from: https://www.nature.com/articles/s41467-019-13036-1.
Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593–8.
McDonald BA, Suffert F, Bernasconi A, Mikaberidze A. How large and diverse are field populations of fungal plant pathogens? The case of Zymoseptoria tritici. Evol Appl. 2022 [cited 2023 Jul 12];15:1360. Available from: https://www.pmc/articles/PMC9488677/.
Ebert D, Fields PD. Host-parasite co-evolution and its genomic signature. Nat Rev Genet. 2020; Available from: http://www.ncbi.nlm.nih.gov/pubmed/32860017.
Lucas JA, Hawkins NJ, Fraaije BA. The evolution of fungicide resistance. Adv Appl Microbiol. Elsevier Ltd; 2015. Available from: https://doi.org/10.1016/bs.aambs.2014.09.001.
Tack AJM, Thrall PH, Barrett LG, Burdon JJ, Laine AL. Variation in infectivity and aggressiveness in space and time in wild host–pathogen systems: causes and consequences. J Evol Biol. 2012 [cited 2022 Jun 6];25:1918–36. Available from: https://onlinelibrary.wiley.com/doi/full/. https://doi.org/10.1111/j.1420-9101.2012.02588.x.
Barrett LG, Thrall PH, Burdon JJ, Linde CC. Life history determines genetic structure and evolutionary potential of host-parasite interactions. Trends Ecol Evol. 2008 [cited 2022 Jun 6];23:678–85. Available from: https://pubmed.ncbi.nlm.nih.gov/18947899/.
New FN, Brito IL. What Is Metagenomics Teaching Us, and What Is Missed? Annu Rev Microbiol. 2020 [cited 2022 May 25];74:117–35. Available from: https://pubmed.ncbi.nlm.nih.gov/32603623/.
Tkacz A, Hortala M, Poole PS. Absolute quantitation of microbiota abundance in environmental samples. Microbiom. 2018 [cited 2024 Jan 9];6:1–13. Available from: https://link.springer.com/articles/. https://doi.org/10.1186/s40168-018-0491-7.
Anderson AJ, Kim YC. Insights into plant-beneficial traits of probiotic Pseudomonas chlororaphis isolates. J Med Microbiol. 2020;69:361–71.
Sánchez-Vallet A, Fouché S, Fudal I, Hartmann FE, Soyer JL, Tellier A, et al. The Genome Biology of Effector Gene Evolution in Filamentous Plant Pathogens.Annu Rev Phytopathol. 2018 [cited 2022 Apr 22];56:21–40. Available from: https://www.annualreviews.org/doi/abs/. https://doi.org/10.1146/annurev-phyto-080516-035303.
Plissonneau C, Hartmann FE, Croll D. Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome. BMC Biol. 2018;16:1–16.
Röttjers L, Faust K. From hairballs to hypotheses–biological insights from microbial networks. FEMS Microbiol Rev. 2018 [cited 2024 Jun 4];42:761–80. Available from: https://doi.org/10.1093/femsre/fuy030.
Araújo MB, Rozenfeld A. The geographic scaling of biotic interactions. Ecography. 2014 [cited 2024 Jun 3];37:406–15. Available from: https://onlinelibrary.wiley.com/doi/full/. https://doi.org/10.1111/j.1600-0587.2013.00643.x.
Pinto S, Benincà E, van Nes EH, Scheffer M, Bogaards JA. Species abundance correlations carry limited information about microbial network interactions. PLoS Comput Biol. 2022 [cited 2024 Jun 5];18:e1010491. Available from: https://journals.plos.org/ploscompbiol/article?id=. https://doi.org/10.1371/journal.pcbi.1010491.
Yeh YC, McNichol J, Needham DM, Fichot EB, Berdjeb L, Fuhrman JA. Comprehensive single-PCR 16S and 18S rRNA community analysis validated with mock communities, and estimation of sequencing bias against 18S. Environ Microbiol. 2021 [cited 2022 May 24];23:3240–50. Available from: https://onlinelibrary.wiley.com/doi/full/. https://doi.org/10.1111/1462-2920.15553.
Wear EK, Wilbanks EG, Nelson CE, Carlson CA. Primer selection impacts specific population abundances but not community dynamics in a monthly time‐series 16S rRNA gene amplicon analysis of coastal marine bacterioplankton. Environ Microbiol. 2018 [cited 2022 May 24];20:2709. Available from: https://www.pmc/articles/PMC6175402/.
Brockhurst MA, Harrison E, Hall JPJ, Richards T, McNally A, MacLean C. The ecology and evolution of pangenomes. Curr Biol. 2019 [cited 2023 Jul 15];29:R1094–103. Available from: https://pubmed.ncbi.nlm.nih.gov/31639358/.
Cummins EA, Hall RJ, McInerney JO, McNally A. Prokaryote pangenomes are dynamic entities. Curr Opin Microbiol. 2022 [cited 2023 Jul 15];66:73–8. Available from: https://pubmed.ncbi.nlm.nih.gov/35104691/.
Tettelin H, Medini D. The Pangenome. The pangenome: diversity, dynamics and evolution of genome. 2020 [cited 2023 Jul 15];1–307. Available from: https://www.ncbi.nlm.nih.gov/books/NBK558825/.
Kiepas, AB, Hoskisson, PA, & Pritchard, L. 16S rRNA phylogeny and clustering is not a reliable proxy for genome-based taxonomy in Streptomyces. Microb Genom. 2024;10(9). Available from: https://doi.org/10.1099/mgen.0.001287.
Rajkumari J, Katiyar P, Dheeman S, Pandey P, Maheshwari DK. The changing paradigm of rhizobial taxonomy and its systematic growth upto postgenomic technologies. World Journal of Microbiology and Biotechnology 2022 38:11. 2022 [cited 2024 Feb 19];38:1–23. Available from: https://link.springer.com/article/. https://doi.org/10.1007/s11274-022-03370-w.
Miranda-Sánchez F, Rivera J, Vinuesa P. Diversity patterns of Rhizobiaceae communities inhabiting soils, root surfaces and nodules reveal a strong selection of rhizobial partners by legumes. Environ Microbiol. 2016 [cited 2024 Feb 19];18:2375–91. Available from: https://onlinelibrary.wiley.com/doi/full/. https://doi.org/10.1111/1462-2920.13061.
Yang L-L, Jiang Z, Li Y, Wang E-T, Zhi X-Y. Plasmids related to the symbiotic nitrogen fixation are not only cooperated functionally but also may have evolved over a time span in family Rhizobiaceae. GBE. 2020 [cited 2023 Sep 29]; Available from: https://jgi.doe.gov/user-programs/.
Barber AE, Sae-Ong T, Kang K, Seelbinder B, Li J, Walther G, et al. Aspergillus fumigatus pan-genome analysis identifies genetic variants associated with human infection. Nat Microbiol . 2021 [cited 2023 Oct 12];6:1526–36. Available from: https://doi.org/10.1038/s41564-021-00993-x.
Kowalski CH, Beattie SR, Fuller KK, McGurk EA, Tang YW, Hohl TM, et al. Heterogeneity among isolates reveals that fitness in low oxygen correlates with Aspergillus fumigatus virulence. mBio. 2016 [cited 2024 Feb 19];7. Available from: https://pubmed.ncbi.nlm.nih.gov/27651366/.
Usyk M, Peters BA, Karthikeyan S, McDonald D, Sollecito CC, Vazquez-Baeza Y, et al. Comprehensive evaluation of shotgun metagenomics, amplicon sequencing, and harmonization of these platforms for epidemiological studies. Cell Reports Methods. 2023 [cited 2024 Feb 19];3:100391. Available from: https://www.pmc/articles/PMC9939430/.