Phylogenomic analyses reveal novel deep-sea fish relationships
Our primary nuclear exon-based phylogenomic dataset included 60 stomiiform species [31 genera, 4 families] and four outgroup taxa, comprising 936 loci totaling 314,607 base pairs (bp), with 73% missing data and a mean of 20 species per locus. To address potential biases from missing data and assess phylogenetic uncertainty, we applied stringent filtering strategies, generating a highly filtered dataset (37 loci: 10,872 bp, 49% missing data), two medium-filtered datasets (401 loci: 160,632 bp, 62% missing data; 472 loci: 223,738 bp, 71% missing sites), and three subset datasets (each 318 loci: 103,515–109,623 nucleotides with 72.14%−73.68% missing sites) (Additional file 1). Maximum likelihood (ML) analyses in IQ-TREE yielded congruent topologies across all datasets, with 78% of nodes in all our trees supported by bootstrap values (BS) > 85%. By contrast, coalescent-based species trees (ASTRAL-IV), inferred from individual gene trees, exhibited significantly lower support values, sharing only 54.4% of nodes on average with concatenated ML trees. Despite substantial discordance in branching patterns between coalescent- and concatenation-based trees, the monophyly of most taxonomic groups proposed herein was consistently resolved (Fig. S1). An exception was Sternoptychidae, which was resolved as paraphyletic in all the coalescent-based analyses, but strongly supported as monophyletic in all concatenation-based analyses (Fig. 2; Fig. S1). The phylogenetic incongruences observed in coalescent-based trees are likely due to the high levels of gene tree estimation error. Given the documented gene tree error and the robust topological consistency across our concatenation-based datasets, we selected the ML tree, inferred from the full 936-locus dataset, as our primary phylogenetic hypothesis. Trees estimated using ASTRAL-IV are summarized in Fig. S1.
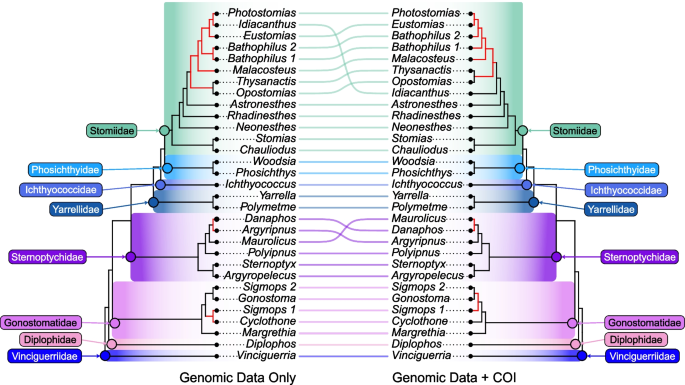
Tanglegram showing concatenation-based stomiiform topologies generated by genomic data only (left) and genomic data supplemented with COI sequences (right). Non-monophyletic genera (Bathophilus & Sigmops) represented as distinct clades (e.g. Bathophilus 1 and Bathophilus 2). Topological discordance indicated with red branches. Family-level classifications follow the new classification system proposed herein
Quality control of COI sequences from BOLD
To assess the reliability of publicly available data for phylogenetic analyses, we assembled a COI dataset by mining 2,474 sequences from the BOLD database and integrating them with COI data from 51 of the 60 species in our primary genomic dataset, resulting in 2,525 individuals with sequence lengths ranging from 567 to 1,563 bp (see Methods). We evaluated sequence quality by inferring an ML tree, followed by visual inspection of the phylogeny for species clustering, topological accuracy, and sequence identification. Sequences were grouped into 181 species-level bins based on consistent clustering (Additional file 2). Comparison of BOLD species names with their bin assignments revealed that 728 sequences (28.8%) failed our quality control (QC) protocol (Fig. S2). Of these, 146 sequences could not be assigned to any bin and were flagged as ‘ID cannot be determined,’ 424 lacked specific genus or species designations (e.g., Stomiiformes sp., Stomias sp.), and 158 showed mismatched bin assignments, indicating misidentification (Fig. S3; Additional file 3). The distribution of QC failures across families revealed that 20.9% of sequences from Gonostomatidae (99/468), 37.1% of Phosichthyidae (130/350), 33.7% of Sternoptychidae (236/701), and 26.3% of Stomiidae (263/998) were problematic (Fig. S3). Overall, 29% of the BOLD sequences (717/2,474) failed our QC protocol. However, genus-level clustering remained largely robust, indicating that inherent challenges associated with identification in this order are concentrated at the species level. To enhance data reliability, we recommend combining automated filtering based on sequence quality metrics with manual verification through preliminary phylogenetic analyses before final tree reconstruction.
Phylogenetic analysis of an expanded multi-locus dataset
To maximize taxonomic coverage, we generated an expanded dataset comprising 135 stomiiform species and four outgroup taxa. We integrated our 936 nuclear loci data matrix (314,607 bp, 73% missing sites) with the COI locus (1,746 bp post-alignment), totaling 316,353 bp with an average of 88% missing sites and a mean of 16 species per locus (Additional file 1). ML analyses based on our expanded dataset in IQ-TREE yielded topologies that retained all major clade relationships from our primary 60-species phylogeny (Fig. 2). As a result of the high levels of missing data, driven by COI-only taxa, and trimmed COI alignment, this reduced resolution at several nodes and required extensive quality control steps. For instance, bootstrap support for Gonostomatidae (as defined herein) decreased from 94% in the primary dataset to < 55% in the expanded dataset. Within Gonostomatidae, the genus Cyclothone exhibited significant phylogenetic uncertainty, resolving within the family in only five of ten ML replicates. In other instances, we resolved a peculiar sister relationship between the gonostomatid genus Cyclothone and the phosichthyid genus Vinciguerria, however, this was likely due to long-branch attraction and missing data. Consistent with previous analyses, several stomiid genera (e.g., Astronesthes, Thysanactis, Photostomias, and Photonectes) have also had unstable placements within Stomiidae. This instability, compounded by variations in taxon sampling and proportionately high levels of missing data, underscores the challenges of resolving phylogenetic relationships with taxonomically expanded datasets, particularly for problematic clades like Cyclothone and the more speciose stomiid genera, such as Astronesthes and Photonectes. Even so, the resolution of higher-level clades were well preserved across most of our analyses (Fig. S1), especially our primary phylogeny (Fig. 2).
Impact of sequence contamination on phylogenetic reconstruction
Given the high rate of contamination identified in the COI dataset, we assessed its impact on phylogenetic inference by reconstructing an ML tree using the 1,797 sequences that passed QC, excluding the 728 problematic sequences (Fig. S3). Compared to the full COI dataset, the filtered dataset improved topological stability for Gonostomatidae, with Cyclothone consistently resolving within the family (BS = 82%) rather than grouping with Vinciguerria as observed in some unfiltered analyses. However, stomiid subfamilies, including Astronesthinae and Stomiinae, continued to exhibit variable support (BS values ranging from 55 to 75%), suggesting that contamination is only one of several factors affecting fine-scale resolution in deep-sea Stomiiformes. A reanalysis of family-level clustering with the filtered dataset also showed improved support for newly proposed families within Stomiiformes, though some inconsistencies persisted compared to past molecular hypotheses (Table 1; Additional file 3). These findings emphasize the critical role of quality control in enhancing phylogenetic accuracy.
Table 1 Assessment of family-level relationships within Stomiiformes using concatenation-based (IQ-TREE) and species tree (ASTRAL-IV & weighted-ASTRAL) methods across 15 datasets. Novel family-level clade comparison across different datasets using concatenation-based and multi-species coalescent methods. The right column includes all Stomiiform families as delimited in this study as well as Smith et al. [13]. Additionally, all previously acknowledged subfamilies of Stomiidae are included. Checkmarks indicate monophyly support for each family or subfamily. Families recognized herein are Vinciguerriidae, Diplophidae, Gonostomatidae, Sternoptychidae, Yarrellidae, Ichthyococcidae, Phosichthyidae, and Stomiidae, with previously-recognized Stomiidae subfamilies being: Stomiinae, Chauliodontinae, Idiacanthinae, Astronesthinae, Melanostomiinae, and Malacosteinae. Families sensu Smith et al. [13] are Gonostomatidae, Sternoptychidae, and StomiidaeNon-monophyly of traditional families reveals complex stomiiform relationships
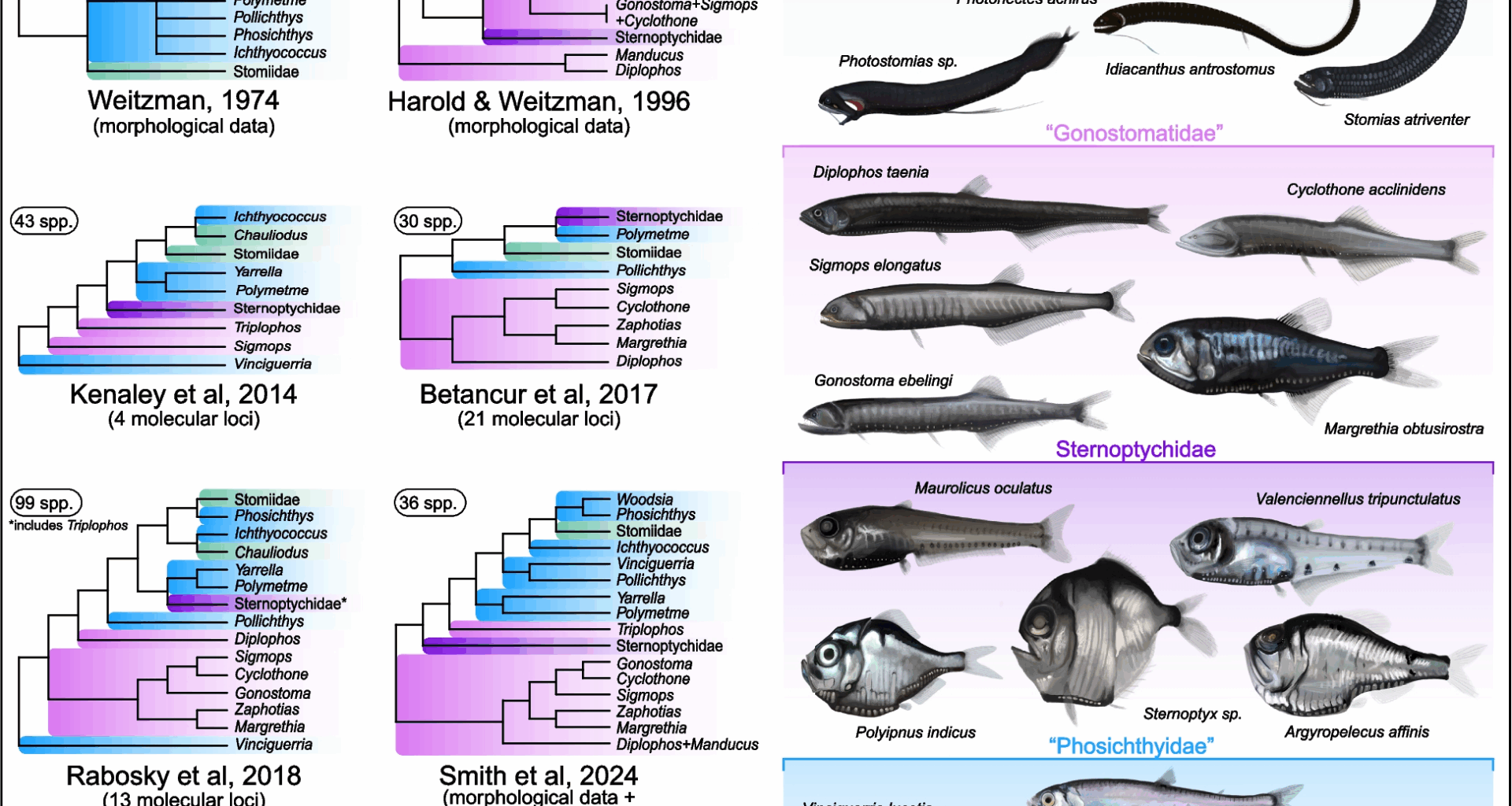
Our analyses fundamentally challenge traditional stomiiform taxonomy by revealing extensive paraphyly and polyphyly in two major families, Gonostomatidae and Phosichthyidae, respectively (as seen in Fig. 3a, b). The gonostomatids were resolved as two separate lineages, with Diplophos forming the sister group to all other taxa except the phosichthyid genus Vinciguerria, which diverges at the base of Stomiiformes (100% BS) (Fig. 3a). The remaining gonostomatid genera (Cyclothone, Gonostoma, Margrethia, Sigmops, Triplophos, and Zaphotias) (Fig. 3b) constitute a robustly-resolved Gonostomatidae sensu stricto, but its intrafamilial relationships are weakly supported and sensitive to changes in species sampling. Phosichthyidae is even more fragmented, with its seven genera scattered across four distinct lineages in nearly all the concatenation- and coalescent-based species trees (Table 1). Even after excluding the early-branching Vinciguerria, the remaining genera form three separate clades, rendering Phosichthyidae highly polyphyletic. One of the remaining three clades, hereafter referred to as Yarrellidae, unites Polymetme with Yarrella as the sister group to the other two remaining phosichthyid groups and the family Stomiidae. In our analyses, Ichthyococcus consistently emerged as an isolated lineage (hereafter, Ichthyococcidae) positioned between Yarrellidae and the newly defined Phosichthyidae (Table 2), now restricted to Phosichthys and Woodsia (Fig. 3a, b). Of the phosichthyid clades, the sole genus Vinciguerria was substantially supported as sister to all other stomiiforms, and is relegated to the new family Vinciguerriidae (along with the morphologically-similar Pollichthys).
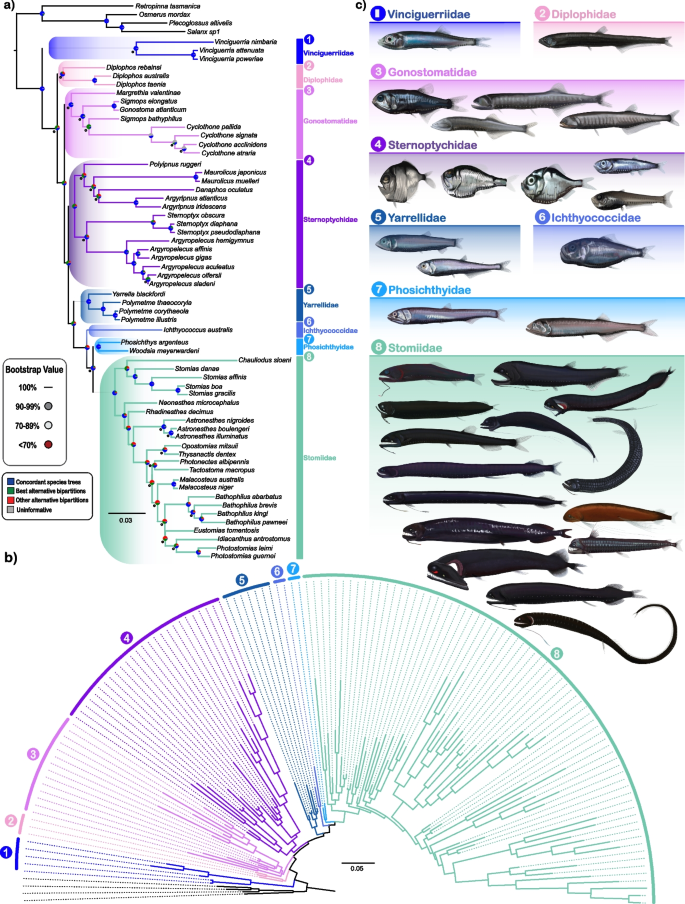
Family-level relationships and morphological diversity of Stomiiformes. a Molecular phylogeny derived from concatenation-based ML analysis of 936 exon markers, illustrating newly proposed family-level relationships. Monophyletic groups are color-coded by family. Nodes with bootstrap (BS) support < 100% are indicated by colored circles (90% and 70% thresholds), in addition to pie charts showing the proportion of concordant trees (blue), best alternative bipartitions (green), other alternative bipartitions (red), and uninformative trees (grey). All trees seen in Table 1 were included in the calculation of concordance of the 60 species phylogeny aside from the ASTRAL subsets. Exact BS values are provided in supplementary files. Numbers, color-coded by family, correspond to morphological illustrations in panel (c). b Radial representation of an expanded (135-taxon) phylogenetic tree based on COI + 936 exon markers, with branch lengths scaled to genetic divergence (scale bar = 0.05 substitutions per site). c Morphological illustrations of representative species from 31 genera across the families (1) Vinciguerriidae, (2) Diplophidae, (3) Gonostomatidae, (4) Sternoptychidae, (5) Yarrellidae, (6) Ichthyococcidae, (7) Phosichthyidae, and (8) Stomiidae, highlighting the morphological diversity within these groups. Families are consistently color-coded across all panels. Illustrations by Casey Lee
Table 2 Systematic classification of Stomiiformes genera: traditional and newly proposed family-level groupings. Comparison of traditional family-level groupings from Fink (1985), Kenaley et al. (2014), and Smith et al. [13] with our newly proposed taxonomic arrangement based on phylogenomic evidence. Relevant stomiiform genera are denoted in the left column while their identified families based on different studies are listed coincidingly including this study. Our new taxonomic classification introduces novel families, including Ichthyococcidae fam. nov., Vinciguerriidae fam. nov., Yarrellidae fam. nov., and a newly revised Phosichthyidae and Gonostomatidae. Dashes (–) indicate missing genera from the study in question. Asterisks (*) highlight genera that have previously been placed in families such as Melanostomiidae, Malacosteidae, and Astronesthidae, which have now been reclassified based on modern phylogenomics.Stomiidae monophyly and Sternoptychidae conflict
Our analyses revealed robust support for a monophyletic Stomiidae (100% BS) (Fig. 3a, b) across all concatenated and most coalescent analyses (Table 1), offering new insights into the evolution of dragonfishes. This well-supported clade encompasses all six traditionally recognized subfamilies, exhibiting moderate to high levels of phylogenetic concordance with minimal conflicting bipartitions when comparing all our analyses to our most stable phylogeny, as indicated by the pie charts in Fig. 3a. However, their internal relationships within Stomiidae challenge existing classifications. Most notably, all three subfamilies containing multiple genera were resolved as polyphyletic (Malacosteinae and Melanostomiinae) or paraphyletic (Astronesthinae). Among these subfamilial groupings, Stomiinae was resolved as monophyletic (Table 1), most often forming a sister clade to a monophyletic Chauliodontinae. However, this latter relationship should be interpreted cautiously, as Chauliodontinae and Idiacanthinae were each represented by a single species in our primary dataset. Given the pervasive non-monophyly observed at both genus and subfamily levels in our analyses, consistent with findings from previous studies, we propose discontinuing the current subfamilial classification within Stomiidae. These artificial groupings appear to obscure, rather than clarify, our understanding of evolutionary relationships among dragonfishes, and their continued use may impede accurate systematic interpretation of this remarkable deep-sea radiation. The marine hatchetfishes (Sternoptychidae) showed strong support for monophyly in concatenated analyses (100% BS) (Fig. 3a) but yielded conflicting signals in species tree analyses (Table 1), which split the family in two distinct groups: Sternoptyx + Argyropelecus and Argyripnus + Maurolicus + Polyipnus + Danaphos. This similarly violates the traditional division of Sternoptychidae into two subfamilies (Sternoptychinae and Maurolicinae), and as such, we do not acknowledge the use of the sternoptychid subfamily classification. Additionally, the position of Sternoptychidae within Stomiiformes fluctuated across analyses (Table 1), indicating that increased taxonomic sampling may be necessary to resolve their phylogenetic position with greater confidence.
Tree concordance factors reveal clade stability and conflict
To assess topological stability while avoiding the biases introduced by gene-tree error, we calculated tree concordance factors (tCFs), which measure the percentage of species trees that resolve each branching pattern in our main phylogeny. The tCFs was computed by using the 13 independently inferred phylogenies, including six concatenated ML trees and seven coalescent species trees and visualized as pie charts showing the proportion of trees supporting each branch (Fig. 3a). In this approach, each inferred tree is treated as a “locus,” adapting the conceptual framework of gene concordance factors (gCFs) to derive tCFs. We observed uniformly high tCFs for several clades defined in our study, Vinciguerriidae, Yarrellidae, and Phosichthyidae each reached 100% concordance, while Gonostomatidae attained 92%. Our revised Stomiidae exhibited 77% concordant splits (23% in the primary alternative), and Diplophidae showed 62% concordance (38% alternative). Notably, Sternoptychidae, despite strong bootstrap support under concatenation, displayed only 38% concordance, reflecting its consistent paraphyly in coalescent‐based trees (Fig. S1). By integrating the signal across methodological and dataset variations, tCFs provide a robust, quantitative measure of both support and conflict in our novel eight-family classification.
Topology testing against alternative classifications
To evaluate the robustness of our proposed eight-family classification against the recently proposed three-family framework by Smith et al. [13]. We conducted Approximately Unbiased (AU) topology tests in IQ-TREE using the 936-locus alignment for both the 64-taxon and 135-taxon datasets. Although our dataset does not statistically reject the monophyly of Stomiidae sensu Smith et al. [13] (p > 0.05), this topology was only ever resolved in one of the 13 species trees (six concatenation-based ML and seven coalescent-based) inferred in our study (Table 1; Fig. S1). Recognizing a monophyletic Stomiidae sensu Smith et al. [13] in our trees would collapse most Stomiiform genera into a single, overly broad Stomiidae. In contrast, our eight-family classification would be more informative and robust, even under topological uncertainty, with high tree concordance factors (tCFs) for most families (e.g., 100% for Vinciguerriidae, Yarrellidae, and Phosichthyidae; 92% for Gonostomatidae) (Fig. 3a). Furthermore, when our eight families are mapped onto the ultraconserved element (UCE) and morphological tree of Smith et al. [13], seven of our eight families remain monophyletic, providing additional evidence of their stability and robustness to topological variation.
Proposed taxonomic classification
Our current classification framework integrates molecular and morphological data to establish a robust taxonomy. We compiled a morphological character matrix (Fig. S4) using traits from Harold & Weitzman (1996) [15], with further validation for some [13, 16, 17, 19]. These traits were mapped onto the ML phylogenetic tree derived from our genomic data (Fig. 3a). We then applied ACCTRAN and DELTRAN character optimizations methods to infer ancestral states on the tree. ACCTRAN prioritizes changes closer to the root, favoring early trait gains and subsequent reversals, whereas DELTRAN shifts changes toward the tips, emphasizing parallel trait evolution (Fig. S5).
Family Diplophidae Fowler 1925
Type genus:Diplophos Günther 1873
Diversity. 9 species in 2 genera.
Diagnosis. Diplophidae is unique among stomiiform families in possessing photophore rows on the lower jaw and lacking pleural ribs associated with the third vertebra. It is further diagnosed by a high vertebral count (44–94 in diplophids vs. 28–60 in other stomiiforms) and a high ventral photophotore count (70–115 vs. 19–95); a well-developed median adductor mandibulae divided into two distinct muscles (vs. reduced in all other stomiiforms); a horizontally oriented extensor proprius pelvicus (vs. vertically-oriented in all other stomiiforms); a large nasal opening extending anteriorly to at least the nasal capsule (vs. small in other stomiiformes); a flattened, dentigerous, and horizontally-oriented basihyal (vs. cylindrical, edentate, and vertically-oriented in all other stomiiforms); and a ventrally bifurcated first pharyngobranchial shaft (vs. non-bifurcated in all other stomiiforms) (Additional file 4).
Genera included.Diplophos (7 spp.) and Manducus (2 spp.).
Comments. The subfamily name “Diplophinae” was first proposed to accommodate the “greatly elongate” and “band-like” body plan of the genus Diplophos [26]. Shortly after, this genus was then reclassified as a maurolicid, and then a gonostomatid. Approximately thirty years later, “Manducinae” was similarly proposed to house Manducus and three other genera [27]. However, both subfamilial classifications failed to be considered and Diplophos and Manducus are currently recognized within Gonostomatidae. Subsequent phylogenetic accounts have varied in their treatments of Diplophos and Manducus, with some studies considering both genera as early-branching stomiiforms [17], or even sister to all other stomiiforms. The name “Diplophidae” was first acknowledged in 2004 [28], but remained controversial and was not widely recognized [3]. Our study corroborates previous analyses, with Diplophos sister to all other stomiiforms except Vinciguerria, and acknowledges the family Diplophidae as its own lineage, based on morphological and molecular evidence (more details in Supplementary Material).
English name. Portholefishes.
Family Gonostomatidae Cocco 1838
Type genus:Gonostoma Rafinesque 1810
Diversity. 25 species in 6 genera.
Diagnosis. Gonostomatidae is diagnosed by a unique combination of traits. The family differs from all stomiiform families except Diplophidae in having beta-type photophores (vs. alpha or gamma in other stomiiforms) and in the presence of an accessory neural arch (vs. absent in other stomiiforms). Gonostomatidae differs from Diplophidae by the following characters: absence of photophore rows on the lower jaw (vs. presence in diplophids); presence of pleural ribs associated with the third vertebra (vs. absence in diplophids); reduction of one or both sections of the median adductor mandibulae (vs. well-developed in diplophids); a vertically-oriented extensor proprius pelvicus (vs. horizontally-oriented in diplophids); a small nasal opening not extending to the nasal capsule (vs. a large and anteriorly-extending nasal in diplophids); a cylindrical, edentate, and vertically-oriented basihyal (vs. flattened, dentigerous, and horizontally-oriented in diplophids); and a non-bifurcated first pharyngobranchial shaft (vs. ventrally-bifurcated in diplophids) (Additional file 4).
Genera included.Cyclothone (14 spp.), Gonostoma (2 spp.), Margrethia (2 spp.), Sigmops (5 spp.), Triplophos (1 sp.), and Zaphotias (1 sp.).
Comments. Within the family, morphological and molecular analyses have reliably supported a monophyletic clade composed of Cyclothone, Gonostoma, Margrethia, Sigmops (sometimes as a synonym of Gonostoma) and Zaphotias (previously Bonapartia) [9, 13, 15]. We find Gonostomatidae to include these five genera and the additional genus Triplophos, which has been considered an early-branching gonostomatid [9] or an ally of Diplophos or the phosichthyids [15] owing to its elongate body and a series of unusual osteological characteristics. Molecular studies have been highly variable in their treatment of Triplophos, with various analyses placing it in Sternoptychidae [29], sister to all stomiiforms besides Vinciguerria and the other gonostomatids [8], or sister to the phosichthyid-stomiid clade “Photichthya” [13]. Our study is unique in resolving Triplophos within a monophyletic Gonostomatidae, and sister to Zaphotias. As such, we recognize six genera within Gonostomatidae.
English name. Bristlemouths. Includes also fangjaws (Gonostoma, Sigmops).
Family Ichthyococcidae fam nov.
Type genus (by monotypy):Ichthyococcus Bonaparte 1840
Diversity. 7 species in 1 genus.
Diagnosis. Ichthyococcidae is unique among stomiiform families in lacking an alveolar process, having the maxilla fused to the anterior supramaxilla, and showing reduction of the premaxilla and the apophyses of the first vertebra. It is further defined by a unique combination of characters. The family differs from all other families except Phosichthyidae (sensu present study) and Stomiidae in having the anal-fin origin posterior to dorsal fin (vs. below or anterior to dorsal fin in other stomiiforms); the bases of the posterior four branchiostegal rays crowded together (vs. widely separated); a reduced mesopterygoid (vs. well developed); and an ascending process of the premaxillary symphysis with straight medial surfaces (vs. curved medial surfaces). Ichthyococcidae further differs from Phosichthyidae and Stomiidae in having a well-developed posterior palatine process (vs. significantly reduced); variably-sized medial jaw teeth (vs. mainly large medial jaw teeth); more than six posterior ceratohyal branchiostegal rays; and fewer than 14 branchiostegal photophores (Additional file 4).
Genera included.Ichthyococcus (7 spp.).
Comments. The phylogenetic placement of genus Ichthyococcus has been historically contentious, but has not changed significantly since its incorporation into Weitzman’s “Phosichthyidae” in the late twentieth century [30]. Since then, Ichthyococcus has been recurrently identified as an outlier within the family, due to its stout body and a number of specialized characteristics, including a beaked mouth and complex sensory structures absent in other phosichthyids [15, 30]. Morphological and molecular analyses have consistently identified strong support for a clade containing Ichthyococcus, Phosichthys, Woodsia, and Stomiidae [8, 15, 29]. Our phylogeny produces identical results, with Ichthyococcus sister to the remaining genera and family mentioned above. Based on previous phylogenomic analyses and distinct characteristics, we recognize a new monotypic family, Ichthyococcidae, to accommodate the genus Ichthyococcus (more details in the Supplementary Material).
English name. Fireflyfishes.
Note: Despite morphological divergence from the “true” lightfishes of genera Phosichthys and Woodsia, species of Ichthyococcus have historically also been called “lightfishes.” To distinguish them from the Phosichthyids, the authors propose the new English name “fireflyfishes,” in reference to their small, compact bodies and bright ventral photophores.
Family Phosichthyidae Weitzman 1974
Type genus:Phosichthys Hutton 1872
Diversity. 3 species in 2 genera.
Diagnosis. Phosichthyidae is defined by a unique combination of traits. This family differs from all other stomiiform families except Stomiidae in having a reduced posterior palatine process (vs. well developed in other stomiiforms); mainly large medial jaw teeth (vs. variably sized); six or fewer posterior ceratohyal branchiostegal rays; and 14 or more branchiostegal photophores. Phosichthyidae further differs from Stomiidae in having the first vertebral parapophyses longer than the second (vs. shorter in stomiids); lacking an ossified Baudelot’s ligament (vs. both shrunken and with ossified Baudelot’s ligament in stomiids); and epineurals fused to neural arches for less than half of the body length (vs. more than half in stomiids) (Additional file 4).
Genera included.Phosichthys (1 sp.) and Woodsia (2 spp.).
Comments. The family “Phosichthyidae” was initially used to categorize seven morphologically-divergent gonostomatid genera—Ichthyococcus, Phosichthys, Pollichthys, Polymetme, Vinciguerria, Woodsia, and Yarrella (Table 2)—characterized by an uneven mixture of anatomical traits typical of both stomiid and non-stomiid families [30]. Anatomical similarities between these genera are tenuous, and morphological analyses have resolved the “phosichthyid” genera as representing up to six different clades within Stomiiformes [15]. Molecular studies have strongly supported this potential non-monophyly, with the seven constituent genera being resolved as two [9], three [8, 13], or four [29] clades, depending on taxon sampling size. A sister relationship between the genera Phosichthys and Woodsia is well-supported by both molecular and morphological evidence [8, 13, 29, 30], including herein. Therefore, we redefine Phosichthyidae to contain only two genera, Phosichthys and Woodsia, with no subfamily-level taxa. The remaining five “phosichthyid” genera—Ichthyococcus, Pollichthys, Polymetme, Vinciguerria, and Yarrella—based on morphological and molecular analyses, do not form a clade with Phosichthys and Woodsia, and are therefore acknowledged within other families herein.
English name. Lightfishes.
Family Sternoptychidae Duméril 1805
Type genus:Sternoptyx Hermann 1781
Diversity. 79 species in 10 genera.
Diagnosis. Sternoptychidae is unique among other stomiiform families in having a single epural; fusion of the third and fourth hypurals; alpha-type photophores; and shortened, subequal parapophyses of the first two vertebrae without an ossified Baudelot’s ligament. The family is further defined by a unique combination of characters. Sternoptychidae shares with some gonostomatids a lateral adductor mandibulae subdivided into dorsal and ventral sections (vs. undivided in other stomiiforms) and protracted photophore metamorphosis (vs. rapid). Sternoptychidae is distinguishable from these gonostomatids by the presence of an ossified accessory neural arch (Additional file 4).
Genera included.Araiophos (2 spp.), Argyripnus (9 spp.), Argyropelecus (7 spp.), Danaphos (2 spp.), Maurolicus (15 spp.), Polyipnus (34 spp.), Sonoda (2 spp.), Sternoptyx (4 spp.), Thorophos (2 spp.), and Valenciennellus (2 spp.).
Comments. Sternoptychidae has been widely recognized as a distinct and monophyletic clade since 1974. This family was formed by the unification of three deep-bodied (“sternoptychine”) and seven shallow-bodied (“maurolicine”) stomiiform genera into a single taxon. Most molecular studies have supported the monophyly of this pairing, with the exception of some topological flukes (e.g., the exclusion of Valenciennellus or inclusion of Triplophos, neither of which have been replicated in subsequent analyses) [8, 9, 13, 31]. Traditionally, Sternoptychidae has been divided into two subfamilies, Sternoptychinae and Maurolicinae (a third, Polyipninae, was proposed in the nineteenth century [32], but was subsumed into Sternoptychinae). Evidence for this two-subfamily system has been intermittent, parsimony analyses of morphology suggested a paraphyly of Maurolicinae with respect to Sternoptychinae [15], and molecular analyses have often found the opposite (paraphyly of Sternoptychinae with respect to Maurolicinae) [9, 29, 31]. Our study supports the latter scenario, the sternoptychine genus Polyipnus forms a low-support clade with the maurolicine genera, rendering Sternoptychinae paraphyletic. Still, our study is lacking data from three maurolicine genera (Araiophos, Sonoda, and Thorophos), which means the monophyly of Maurolicinae cannot be adequately evaluated. Owing to topological uncertainty across recent studies, and an absence of genetic data for important genera, we recommend the temporary discontinuation of subfamily-level taxa in Sternoptychidae, until advanced techniques with further taxon sampling are able to confidently clarify genus-level relationships. We adopt the traditional ten-genus definition of Sternoptychidae, with no subfamily-level classifications.
English name. Hatchetfishes (Argyropelecus, Polyipnus, Sternoptyx); pearlsides (Araiophos, Argyripnus, Maurolicus, Sonoda, Thorophos); bottlelights (Danaphos); and constellationfishes (Valenciennellus).
Family Stomiidae Bleeker 1859
Type genus:Stomias Jordan & Seale 1906
Diversity. 327 species in 27 genera.
Diagnosis. Stomiidae is unique among stomiiform families in having an ossified Baudelot’s ligament on the first two vertebral parapophyses; lacking gill rakers in adults; absence of the sixth hypural; division of the geniohyoideus into dorsal and ventral portions; and anterior and posterior enlargement of the posterior pelvic plate, with a cartilaginous core extending posteriorly beyond the ossified portion of the plate (Additional file 4).
Genera included.Aristostomias (6 spp.), Astronesthes (50 spp.), Bathophilus (20 spp.), Borostomias (6 spp.), Chauliodus (9 spp.), Chirostomias (1 sp.), Echiostoma (1 sp.), Eupogonesthes (1 sp.), Eustomias (134 spp.), Flagellostomias (1 sp.), Grammatostomias (4 spp.), Heterophotus (1 sp.), Idiacanthus (3 spp.), Leptostomias (12 spp.), Malacosteus (2 spp.), Melanostomias (18 spp.), Neonesthes (2 spp.), Odontostomias (2 spp.), Opostomias (2 spp.), Pachystomias (1 sp.), Photonectes (29 spp.), Photostomias (6 spp.), Rhadinesthes (1 sp.), Stomias (12 spp.), Tactostoma (1 sp.), Thysanactis (1 sp.), and Trigonolampa (1 sp.).
Comments. The historical delimitations of family Stomiidae have been largely uncontroversial for forty years, since the demotion of six families (Astronesthidae, Chauliodontidae, Idiacanthidae, Malacosteidae, Melanostomiidae, and Stomiidae) into subfamilies within Stomiidae [16]. Early molecular investigations have occasionally failed to resolve a monophyletic Stomiidae due to the exclusion of the subfamily Chauliodontinae [8, 29]. However, phylogenomic investigations, present study included, reliably recover a monophyletic and well-supported Stomiidae that includes the chauliodontines [9, 13]. Unlike other studies [8, 16], our results discourage the use of these subfamily-level classifications. The astronesthines are split across three lineages, the malacosteines across three more, and the melanostomiines across seven. The three remaining subfamilies — Chauliodontinae, Idiacanthinae, and Stomiinae — resolve as monophyletic in our phylogeny by virtue of monotypy. Due to the topological uncertainty and lack of resolution of the six-subfamily system across multiple studies, including herein, we recommend the recognition of a monophyletic Stomiidae, containing its 27 traditional genera, but we also disregard the Stomiidae sub-family system until adequate taxonomic sampling and consistent placement of stomiids within the family have been resolved (more details in the Supplementary Material).
English name. Dragonfishes. Includes also stareaters (Astronesthes); loosejaws (Aristostomias, Grammatostomias, Malacosteus, Photostomias); boafishes (Stomias); snaggletooths (Borostomias, Eupogonesthes, Heterophotus, Neonesthes, Rhadinesthes); viperfishes (Chauliodus); and sawtails (Idiacanthus).
Family Vinciguerriidae fam. nov.
Type genus:Vinciguerria Jordan & Evermann 1896
Diversity. 6 species in 2 genera.
Diagnosis. Vinciguerriidae is unique among other stomiiform families in having an elongate hyomandibular spine bound to the surface of the mesopterygoid by a ligament (vs. short and detached from mesopterygoid in all other stomiiforms); fusion or tight adherence of the second basibranchial tooth plates (vs. absent or loosely adhering in other stomiiforms); and close adherence of the third basibranchial tooth plates on the dorsal surface (vs. lateral to basibranchial). The family is further defined by a unique combination of traits. Vinciguerriidae differs from the other early-branching families (Diplophidae, Gonostomatidae, and Sternoptychidae) by presence of a posterior photophore; gamma-type photophores; an anteriorly angled dorsal uncinate process of the second pharyngobranchial (vs. straight and vertical in aforementioned families); and presence of a serial photophore duct and lumen.
Vinciguerriidae differs from Yarrellidae in having a posterior photophore; a-cell radiating configuration (vs. irregular in yarrellids); separate contralateral and ipsilateral branches of the premaxillary-rostrodermethmoid ligament (vs. fused in yarrellids); and presence of tooth plates on the fourth basibranchial. Lastly, Vinciguerriidae differs from the remaining families (Ichthyococcidae, Phosichthyidae, and Stomiidae) in having the anal fin originating below or anterior to the dorsal-fin origin (vs. posterior in aforementioned families); the posterior four branchiostegal separated from one another; an anterior palatomaxillary ligament looped over the dorsal surface of the lateral process of the rostrodermethmoid; a well-developed mesopterygoid; a medial concavity of the ascending process of the premaxillary symphysis; and the presence of tooth plates on the fourth basibranchial (Additional file 4).
Genera included.Pollichthys (1 sp.) and Vinciguerria (5 spp.).
Comments.Vinciguerria was initially described as a diminutive “maurolicid” in 1896 [33] but was eventually relocated into Gonostomatidae. In 1959, the morphologically similar Pollichthys was also described as a fellow gonostomatid [34], until both were eventually recognized within Phosichthyidae in 1974 [30]. Since then, their placement has remained contentious, though they reliably exhibit a sister relationship to one another [11, 15]. Further molecular analyses have frequently reaffirmed this relationship, in addition to the Pollichthys + Vinciguerria clade as early-branching within stomiiforms [8, 9, 13, 29]. Although our dataset only incorporated Vinciguerria, based on previous morphological and molecular studies, we are confident in hypothesizing a sister relationship between Vinciguerria and Pollichthys [13, 15]. We relocate both genera to the new family Vinciguerriidae, a morphologically distinct lineage of early-branching stomiiforms that has been reliably resolved across multiple studies.
English name. Lighthousefishes (Vinciguerria) and stareyes (Pollichthys).
Family Yarrellidae fam. nov.
Type genus:Yarrella Goode & Bean 1896
Diversity. 8 species in 2 genera.
Diagnosis. Yarrellidae is unique among other stomiiform families in having the contralateral and ipsilateral branches of the premaxillary-rostrodermethmoid ligament fused into a continuous sheet of connective tissue (vs. unfused in all other stomiiforms). Yarrellidae is further defined by a unique combination of characters. The family differs from Diplophidae, Gonostomatidae, and Sternoptychidae by the presence of a second epural (vs. absence in those families) and by having a gamma-type photophores (vs. alpha or beta). Yarrellidae is distinguishable from the remaining families (Ichthyococcidae, Phosichthyidae, Stomiidae, and Vinciguerriidae) by the absence of a posterior orbital/postorbital photophore (vs. presence in those families) and by the irregular configuration of photophore A cells (vs. regular) (Additional file 4).
Genera included.Polymetme (6 spp.) and Yarrella (2 spp.).
Comments. The genera Yarrella and Polymetme were described in 1896 and 1926, respectively, as gonostomatids. They were relocated into Phosichthyidae in 1974 [30], where they were considered deeply divergent within the family. Morphological analyses subsequently found Polymetme forming a clade with all other phosichthyids and the family Stomiidae, to the exclusion of Yarrella. Past molecular analyses have also resolved a Polymetme + Yarrella sister relationship, which was in turn sister to a clade containing Ichthyococcus, Phosichthys, Woodsia, and the stomiids [8, 13, 15]. The repeated recovery of this clade leads us to support the recognition of a new family, Yarrellidae, for these two divergent genera, previously considered as phosichthyids (more details in Supplementary Material).
English name. Rendezvousfishes.