Newswise — In intercropping systems, soybeans grown beneath tall maize plants experience severe shading that disrupts photosynthesis, weakens stems, and limits yield. Previous genetic studies identified several loci related to stem strength or morphology but failed to explain the complexity of shade adaptation. Shade tolerance involves multiple physiological responses, from signal perception to growth adjustment, suggesting a polygenic basis. However, the connections among these genes and their chronological interactions remained elusive. Based on these challenges, it was necessary to conduct a comprehensive exploration of soybean’s genetic and molecular network underlying shade tolerance through integrated forward and reverse genetic approaches.
A research team from Nanjing Agricultural University and the Guangxi Academy of Agricultural Sciences published their study (DOI: 10.1093/hr/uhae333) on March 1, 2025, in Horticulture Research. The paper presents the first complete model of how soybean genes interact to resist shading. Using advanced GASM-RTM-GWAS and transcriptomic analyses, the team identified key regulatory modules and twelve hub genes that orchestrate the plant’s adaptive response to low-light environments.
To decode soybean’s shade tolerance, the team developed a recombinant inbred line population derived from two contrasting parents—one shade-tolerant and one sensitive. Using the innovative gene–allele sequence-based GWAS (GASM-RTM-GWAS), they identified 211 candidate genes associated with two key indicators: shade tolerance index and relative pith cell length. These genes clustered into five major biological categories, including signal transduction, gene expression, catalysis, primary metabolism, and unknown functions, forming an interconnected protein–protein interaction network.
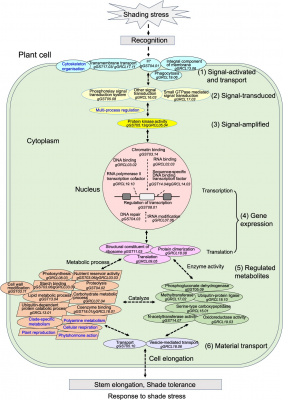
Complementary transcriptomic analysis revealed 7,837 differentially expressed genes (DEGs), 85% of which interacted within large expression networks. Combining both datasets, the team outlined six chronological gene modules—ranging from signal activation to material transport—that describe the plant’s dynamic response to shading. Twelve key genes, such as Glyma.17G136400, Glyma.05G220900, and Glyma.11G003700, were highlighted as central regulators, mediating processes from signal amplification to ribosomal activity. The study demonstrates that soybean shade tolerance emerges from an intricate, multi-stage gene network rather than isolated gene effects.
“Traditional genetics focused on individual genes, but complex traits like shade tolerance require a systems-level perspective,” said Professor Junyi Gai, the study’s corresponding author. “Our integrated approach combines causal gene discovery with expression profiling to visualize the entire regulatory network. This model explains not only how soybeans sense and respond to shading stress but also how multiple genes interact in sequence to maintain growth balance. The identification of key hub genes offers a solid foundation for targeted molecular breeding.”
The comprehensive mapping of the soybean shade tolerance gene network provides valuable insights for future breeding programs. By identifying the hub genes and their regulatory modules, breeders can now target specific stages of the shade response to enhance adaptability in intercropping systems. This framework also establishes a methodological model for studying other complex quantitative traits in crops. Ultimately, integrating genetic, transcriptomic, and functional analyses could accelerate the development of climate-resilient and resource-efficient soybean varieties—boosting productivity in sustainable agriculture and contributing to global food security.
###
References
DOI
Original Source URL
https://doi.org/10.1093/hr/uhae333
Funding information
This work was financially supported by the grants from the National Key Research and Development Program of China (2021YFF1001204, 2021YFD1201602), the MOE 111 Project (B08025), the MOA CARS-04 program, the Zhongshan Biological Breeding Laboratory program (ZSBBL-KY2023-03), the Core Technology Development for Breeding Program of Jiangsu Province (JBGS-2021-014), the Guangxi Scientific Research and Technology Development Plan (14125008-2-16), and the Guidance Foundation of Sanya Institute of NAU (NAUSY-ZZ02, NAUSY-MS05).
About Horticulture Research
Horticulture Research is an open access journal of Nanjing Agricultural University and ranked number one in the Horticulture category of the Journal Citation Reports ™ from Clarivate, 2024. The journal is committed to publishing original research articles, reviews, perspectives, comments, correspondence articles and letters to the editor related to all major horticultural plants and disciplines, including biotechnology, breeding, cellular and molecular biology, evolution, genetics, inter-species interactions, physiology, and the origination and domestication of crops.