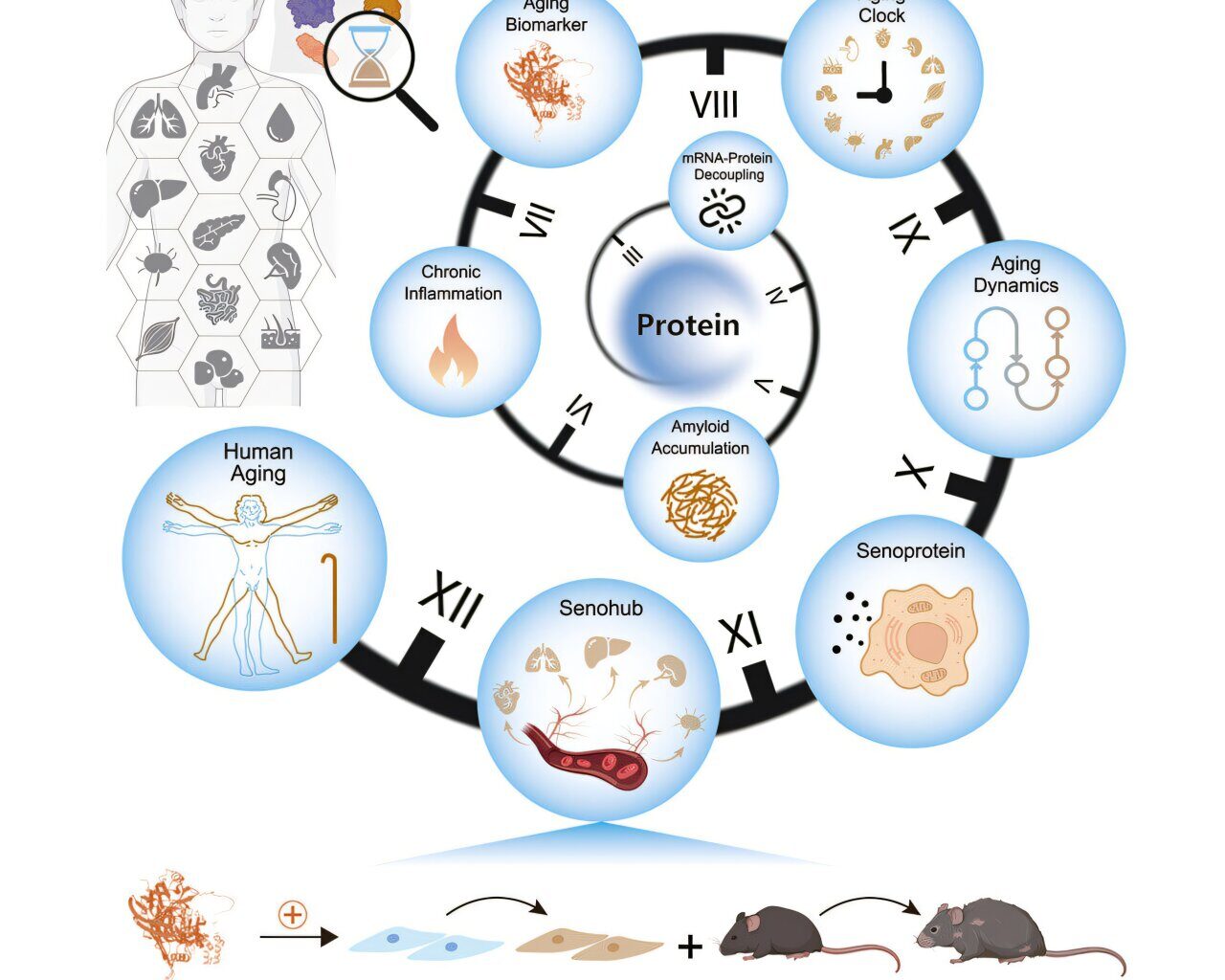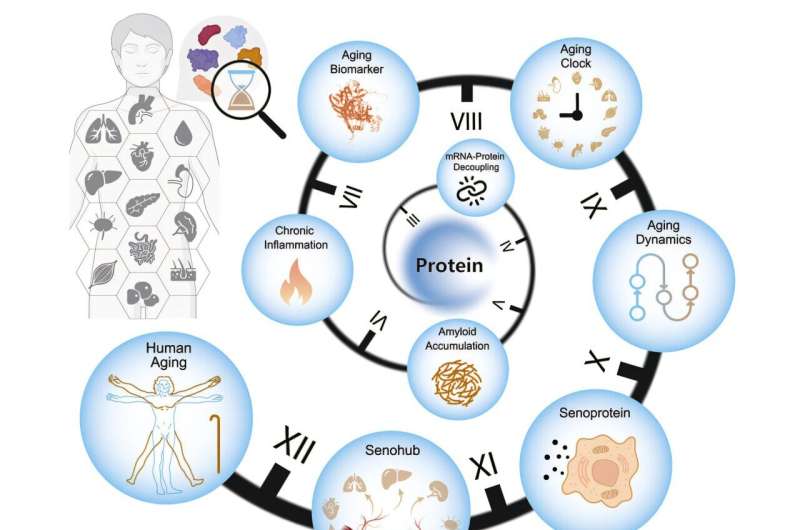
Graphical abstract. Credit: Cell (2025). DOI: 10.1016/j.cell.2025.06.047
A multi-institutional team led by the Chinese Academy of Sciences has constructed a proteomic atlas of human aging across 13 organs, revealing tissue-specific aging clocks, transcriptome-proteome decoupling, and secreted proteins that may accelerate systemic decline.
Organ-specific aging and deterioration drive vulnerability to chronic diseases. Previous studies focused primarily on plasma proteins or DNA methylation profiles. No investigation has systematically mapped how protein quality control deteriorates differently across tissues identified organ‑specific biological age biomarkers.
In the study, “Comprehensive human proteome profiles across a 50-year lifespan reveal aging trajectories and signatures,” published in Cell, researchers designed a multi-tissue proteomic analysis, charting organ-level protein dynamics and aging-related biomarkers across five decades of adult life to construct a longitudinal proteomic atlas of human aging.
Samples from 76 individuals aged 14 to 68, spanning 516 tissue specimens and plasma, were profiled using high-resolution mass spectrometry and parallel transcriptomic assays.
More than 12,700 proteins were quantified across cardiovascular, digestive, immune, endocrine, respiratory, integumentary, and musculoskeletal systems. Subcellular mapping showed that intracellular proteins dominated, particularly those in nuclei and mitochondria. Cross-organ comparisons identified both tissue-enriched signatures and common proteins involved in essential cellular functions.
Age-associated loss of mRNA-protein correlation was observed across all tissues, especially in spleen, lymph nodes, and muscle. Parallel declines were found in protein synthesis, folding, and degradation machinery, including ribosomal subunits and chaperonins.
Amyloid proteins such as SAA1 and SAA2 accumulated along with immunoglobulins and complement factors, forming what the authors describe as an amyloid-immunoglobulin-complement axis within aging tissues. This triad exemplifies how mis‑folded amyloid deposition can trigger immune complex activation and complement‑driven inflammation, linking intracellular quality‑control collapse to tissue-wide issues.
Differentially expressed proteins (DEPs) were grouped into modules by age trajectory and tissue distribution. A set of 29 proteins, including serum amyloid P-component (SAP), rose consistently across at least six tissues.
Another group of 31 proteins, such as PRPF4, showed uniform decline. Histological analysis confirmed SAP accumulation in multiple organs. Functional assays showed that SAP triggered senescence and inflammation in human aortic endothelial cells.
To assess biological age across organs, researchers constructed tissue-specific proteomic clocks using elastic net regression. Predictive performance reached Spearman’s r values between 0.74 and 0.95. TIMP3, a metalloproteinase inhibitor, appeared in nine distinct clocks. Midlife aging inflections emerged between ages 45 and 55, with the aorta showing the most sustained changes.
Secretory protein analysis identified distinct modules of age-responsive factors. CXCL12, a chemokine, accumulated across nine tissues and matched known senescence-associated secretory phenotype (SASP) genes.
Tissue-level ligand-receptor mapping revealed that the aorta, spleen, and adrenal glands maintain extensive cross-talk with other organs. These interactions intensified with age and were mediated by chemokines and adhesion molecules.
Ligand-receptor analysis of plasma proteins pinpointed 24 senescent communication pairs, implicating the aorta as a central initiator of systemic aging—termed a “senohub” by the authors.
One such initiator, GAS6, increased with age in both plasma and aorta and interacted with TAM family receptors. In human endothelial and smooth muscle cells, GAS6 induced senescence markers, elevated IL-6, impaired angiogenic capacity, and promoted immune cell adhesion. In middle-aged mice, intravenous GAS6 injections triggered vascular dysfunction, tissue inflammation, and physical decline.
A total of 211 proteins showed parallel increases in both plasma and at least one tissue. A separate set of proteins, including HARS1 and CCT5, declined with age in both compartments.
To evaluate causal effects, researchers tested seven of the upregulated plasma proteins in human endothelial cells. GPNMB, COMP, HTRA1, SLPI, IGFBP7, NEGR1, and NOTCH3 each induced senescence hallmarks, inflammatory signaling, impaired migration, and angiogenesis deficits. GPNMB administration in mice reduced motor performance, increased vascular inflammation, and induced degeneration in multiple tissues.
Additional observations from the study include widespread epigenetic instability, a coordinated decline in mitochondrial function, remodeling of key protein complexes, and an early‑onset aging signal originating in the adrenal gland.
Researchers conclude that vascular tissues act both as early sensors and as transmitters of aging signals. Proteins secreted by aged vessels may amplify senescence activities throughout the body. Blood-borne factors such as GAS6 and GPNMB function as molecular agents of inter-organ aging.
Written for you by our author Justin Jackson,
edited by Sadie Harley, and fact-checked and reviewed by Robert Egan—this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive.
If this reporting matters to you,
please consider a donation (especially monthly).
You’ll get an ad-free account as a thank-you.
More information:
Yingjie Ding et al, Comprehensive human proteome profiles across a 50-year lifespan reveal aging trajectories and signatures, Cell (2025). DOI: 10.1016/j.cell.2025.06.047
Journal information:
Cell
© 2025 Science X Network
Citation:
Human proteome study maps aging signatures across 13 organs (2025, July 29)
retrieved 29 July 2025
from https://medicalxpress.com/news/2025-07-human-proteome-aging-signatures.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.