WHAT IS ALREADY KNOWN ON THIS TOPICHOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study provides novel insights into the relationship between genetic predisposition to sedentary behaviour and the development of cardiovascular disease, shedding light on a previously underexplored aspect of disease aetiology.
Understanding the contribution of genetics to sedentary behaviour and related health consequences may motivate health professionals to develop novel physical activity interventions.
Introduction
Cardiovascular diseases (CVDs) are the leading cause of disease burden and death worldwide.1 Sedentary behaviour is any waking behaviour in a sitting, reclining or lying posture and characterised by an energy expenditure of less than or equal to 1.5 metabolic equivalents (METs).2 Adults spend up to 60% of their awake time sedentary.3 The prevalence of sedentary behaviours has remained stable or even increased during the last 20 years.4 Sedentary behaviours are systematically associated with increased risk of CVD incidence and related mortality in prospective observational cohort studies.5 6
Interestingly, energy-saving sedentary behaviour has been suggested to be a trait selected by evolution to ensure reproduction and survival.7 Thus, genetics may be an important driver of sedentary behaviour, as supported by previous evidence from twin and candidate gene studies.8 However, the potential effects of this genetic liability on morbidity are unclear. From an evolutionary perspective, it is plausible that genetic predisposition to sedentary behaviour increases the risk of common non-communicable diseases.9 10 It has been suggested that the rapid environmental changes in societies, particularly in Western countries, may have led to an ‘evolutionary mismatch’.9 In this scenario, the previously advantageous alleles, for example, the genetic liability for sedentary behaviour, may cause morbidity in the current postindustrial environment.11 Furthermore, as life expectancy has increased, the later life deleterious effects of underlying genetics may become more apparent.10 The theory of antagonistic pleiotropy suggests that some genes that have favoured survival during reproductive ages in the past may currently limit lifespan and increase morbidity.12 Underlying genetics may, therefore, be an important confounding factor in the commonly observed associations between sedentary behaviour and incident CVD.
Polygenic scores (PGSs) summarise the genome-wide information between single-nucleotide polymorphisms (SNPs) and a phenotype into a single variable that quantifies a person’s genetic liability for a given trait.13 In this study, we assess whether a higher genetic liability for sedentary behaviour is associated with an increased risk of incident CVD (based on individual-level data and measured by hazard ratios and incidence rates). Leisure screen time (LST) is a commonly reported voluntary mode of sedentary behaviour and thus serves as a proxy for this trait.6 We hypothesised that higher genetic liability for LST, as measured by a PGS LST, would be associated with an increased risk of developing CVD.
Methods
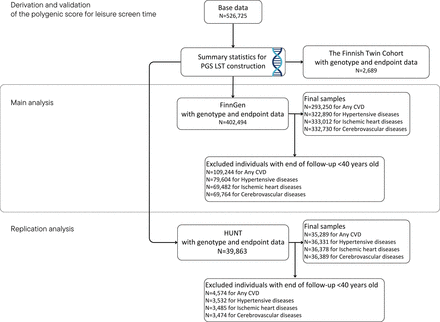
This study consists of four phases. First, we constructed a PGS for LST. Second, we assessed the validity of this score against self-reported LST in the older Finnish Twin Cohort (FTC). Third, we assessed the associations between the PGS LST and incident CVD in the FinnGen cohort (~10% of the Finnish population). Finally, we replicated the analyses in an independent cohort (The Norwegian Trøndelag Health Study (the HUNT study)). The study design and cohorts are shown in figure 1. Additionally, we conducted exploratory analyses to estimate the potential explanations for the observed associations. Detailed information regarding the cohorts is available in the online supplemental methods.
Study design and cohorts. CVD, cardiovascular disease; HUNT, Norwegian Trøndelag Health Study; PGS LST, polygenic score for leisure screen time.
OutcomesLeisure screen time
For the validation analyses in the FTC, LST in hours per day (hour/day) was used as the outcome and included self-reported television (TV) and video viewing, and computer use at home. This outcome was selected to match exactly the outcome used in the base data for PGS LST. In the FTC, participants reported how many hours per day they spent sitting: at home watching TV or videos, and at home at the computer using two separate questions. Both questions had four choices: (a) less than an hour, (b) an hour–less than 2 hours, (c) 2 hours–less than 4 hours and (d) 4 hours or more.14 Answers were summarised to indicate LST (hour/day). The data were collected in 2011 and represent screen time activities at that time.
Cardiovascular disease
Any CVD and the three most common CVDs in the FinnGen, that is, hypertensive, ischaemic heart and cerebrovascular diseases were selected. The CVD categorisation was based on FinnGen endpoint definitions and ICD-10 and ICD-9 codes detailed in the online supplemental methods. We used the same ICD codes in the HUNT replication analyses.
Exposure
The PGS LST was based on the largest available genome-wide association study (GWAS) meta-analysis for LST at the time of the analysis (GWAS Catalogue Study ID: GCST90104339).15 The SNP heritability was 7.4% assessed by Complex-Traits Genetics Virtual Lab.16 The base data did not overlap with the later used FTC, FinnGen or HUNT study cohorts. The descriptives of the base data are described in online supplemental table 1.
We used the high-performing SBayesR approach to construct the PGS.17 In SBayesR, the base data summary statistics are reweighted to consider the linkage disequilibrium (LD) between each variant and restricted to HapMap3 SNPs to ensure computational efficiency while representing the whole genome. The SBayersR method does not use any specific p value threshold nor pruning for selecting the informative variants. The method uses Bayesian multiple regression models, where GWAS SNP effect estimates are scaled using a mixture of four alternative genetic effort distributions as the prior.17 18 The correlations between genetic variants (LD) were taken into account by using an external LD-reference panel generated by the SBayesR authors, constructed from a random sample of 50 000 UK Biobank individuals. These reweighted effect sizes (shown in online supplemental file 2) are then summarised to form a PGS, which indicates the genome-wide genetic liability for the trait of interest. The pipeline and relevant codes are available elsewhere.19 The final number of processed variants was 890 575 in the FTC, 891 628 in the FinnGen cohort and 901 640 in the HUNT cohort. We used standardised PGS LST scores (mean of 0, SD of 1) in the analyses.
Genotyping, quality control and imputation
Details related to genotyping procedures in the different cohorts are available in the online supplemental methods.
Covariates
Covariates were based on previous literature.20 21 The first 10 principal components of ancestry adjust for any genetic stratification that may occur in the study population and were used alongside sex as a covariate in all analyses.20 Additional covariates were specific to the analysis. In validation analyses in the FTC, we also used age, educational attainment (highest achieved self-reported educational degree in the Finnish educational system; low, basic education degree at most; middle, basic education and additional studies; and high, at least upper secondary degree),22 and body mass index (BMI) based on valid self-reported height and weight.23 In main and replication analyses, we also used genotyping batch, which accounts for the potential stratification that may occur from the genotyping array and sample. In further exploratory analyses with the replication cohort, we controlled for variance caused by highly plausible covariates, such as BMI (based on measured height and weight (Jenix DS-102, Dong Sahn Jenix Co, Ltd, Korea)), socioeconomic status (SES; derived from occupational status)21 and self-reported smoking status (never, former, current) using the HUNT3 data.
Statistical analysisValidation analyses
All validation analyses were performed with Stata BE 18.0 (StataCorp). Correlation between PGS LST and self-reported LST was tested with Pearson correlation. The association between PGS LST and self-reported LST was assessed with linear regression in the FTC, where the twin structure of the data was acknowledged using the svyset command (ie, the twin pair was the primary sampling unit). The crude model included standardised PGS LST and the first ten principal components of ancestry. Model 1 was adjusted additionally for age and sex, and model 2 was further adjusted with educational attainment and BMI. Differences between high and low genetic liabilities were evaluated using linear regression, where a binary variable indicated whether the individual belonged to the highest or lowest decile of PGS LST. A priori power calculations indicated that sufficient statistical power would be reached with a sample size of N>950 (expected small effect size 0.02, 15 predictors, power at 0.8 level and α set to 0.05).
Main analyses
We used Cox proportional hazard models and R packages survival24 and survminer25 to estimate the hazard ratios (HRs) and 95% confidence intervals (CI) between PGS LST and incident CVD in the FinnGen cohort. Visual inspection of Schoenfeld residuals and log-minus-log plots indicated that the fully adjusted models satisfy the proportional hazard assumptions.26 PGS×SEX interaction was tested, but no replicable sex differences were observed; thus, analyses were not disaggregated by sex. We used age as the time scale and the first 10 principal components of ancestry, genotyping batch, and sex as covariates. The start of follow-up was set at birth, as the PGS remains stable from conception. The follow-up ended with whichever came first among the first record of the endpoint of interest, death or the end of follow-up on 31 December 2021. Using the weights of the European standard population, age-standardised incidence was calculated when relevant.27 We also compared the HRs (relative measures of survival) and cohort-specific cumulative incidence of events (absolute measures of survival) between the highest and lowest deciles of PGS LST. We additionally conducted competing risk analysis with Fine-Gray subdistribution hazard models for the three main CVDs, as one event may preclude the occurrence of another.28 Additionally, as the exposure (PGS LST) might affect who survives until genotyping and also to control for immortal time bias, we conducted sensitivity analyses in which the start of the follow-up was set to the date of individual DNA sampling. Sufficient statistical power was expected to be achieved with >120 cases, using a classical definition of 10 cases per predictor variable.
Replication analyses
The replication analyses were conducted in HUNT and followed a similar procedure as the main survival analyses, except that when mortality data were not available, the end of follow-up was assigned to whichever came first between the first record of the endpoint of interest and last verified contact with healthcare.
Exploratory analyses
We tested if the associations were independent of highly plausible covariates with a complete case subsample of HUNT3. In this analysis, model 1 included PGS LST, the first 10 principal components of ancestry, sex, and genotyping batch while model 2 included additionally SES, model 3 additionally BMI and model 4 additionally smoking status. We also assessed if adding PGSs for CVDs and physical activity (PA) (online supplemental methods) attenuates the main observations. We additionally tested a potential mediating pathway from PGS LST to incident CVD via sedentary behaviour using the product of coefficients approach and accompanying Sobel test, which are suitable for survival analysis with common outcomes.29 In this mediation analysis, we used participants from the sensitivity analyses with self-reported total sitting time (hour/day): “About how many hours do you sit during an average day? (include work hours and leisure time)”. Only diseases with an established association between PGS LST and incident disease were included in the mediation analysis. The association between PGS LST and total sitting time was evaluated with linear regression, while a Cox proportional hazards model was used for the association between total sitting time and disease incidence. The models were adjusted for sex similar to prior sensitivity analyses but not for the first ten principal components or genotyping batch for consistency across pathways. These mediating analyses were replicated with a PA variable, that is, MET hours per week, calculated as the product of self-reported exercise frequency, intensity and duration as previously described elsewhere.21 30 Across all analyses, all tests were two-tailed, and α was set at p<0.05.
Patient and public involvement
The participants were not involved in forming the research questions, outcome measures, design or other aspects of the study.
Equity, diversity and inclusion statement
The author group is gender-balanced, consisting of five women and six men. The team comprises PhD students, mid-career as well as senior researchers from diverse disciplines. The authors are from two countries. However, both represent high-income countries. Our study population, sampled on a population basis, includes both male and female subjects (52% women) from diverse socioeconomic backgrounds. However, those with access to healthcare might be over-represented in the sample.
ResultsAssociation between PGS for LST and self-reported LST
The 2689 individuals of the FTC reported an average of 3.9±1.1 hours LST per day (online supplemental table 2). The effect size for PGS LST against LST was small (Pearson correlation coefficient=0.08, p<0.001), and PGS LST explained 0.7% of the variance in the LST. PGS LST of 1 SD higher was associated with higher self-reported LST (hour/day), with β=0.09 (95% CI 0.05 to 0.14) in the crude model; and β=0.08 (95% CI 0.04 to 0.13) in the fully adjusted model (online supplemental table 3). Correspondingly, individuals in the highest decile for PGS LST reported an average of 4.1±1.0 hour/day LST, while those in the lowest decile reported an average of 3.7 (1.1) hour/day (p<0.001 for difference) (online supplemental table 4).
Association between PGS for LST and CVD incidence
Descriptive data on 293 250 participants in the FinnGen and 35 289 participants in the HUNT with any CVD are shown in table 1. The average follow-up time from birth was 58 (40 to 106) years (total of 17 101 133 person-years) in FinnGen and 64 (40 to 103) years (total of 2 257 493 person-years) in the HUNT.
Table 1
Descriptives of individuals with any CVD in the main FinnGen cohort and in the replication cohort (HUNT)
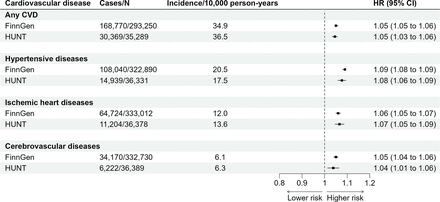
In FinnGen, PGS LST of 1 SD higher was associated with a higher risk of any CVD after adjusting for the first 10 principal components of ancestry, sex and genotyping batch (HR 1.05, 95% CI 1.05 to 1.06; figure 2). Disease-specific associations for the three most common CVDs were (1) 1.09 (95% CI 1.08 to 1.09) for hypertensive diseases (108 040 cases), (2) 1.06 (95% CI 1.05 to 1.07) for ischaemic heart diseases (64 724 cases) and (3) 1.05 (95% CI 1.04 to 1.06) for cerebrovascular diseases (34 170 cases). The associations were replicated in the HUNT cohort with exactly or very similar effect sizes (figure 2).
Associations between polygenic score for leisure screen time and CVD incidence. Values are HRs and 95% CIs per 1 SD increase in the polygenic score for leisure screen time. We used age as the time scale, and the first 10 principal components of ancestry, sex and genotyping batch as covariates. Incidence values are age-standardised according to the European Standard Population to enable comparison between FinnGen and HUNT cohorts. CVD, cardiovascular disease; HUNT, Norwegian Trøndelag Health Study.
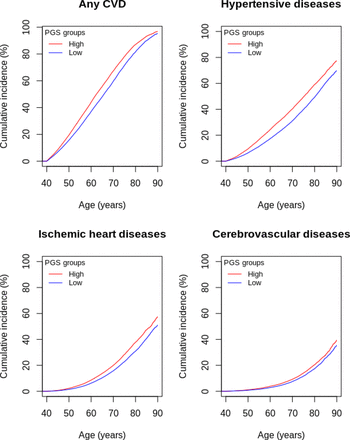
In FinnGen, those in the highest PGS LST decile had 19%–35% higher risk of CVDs compared with those in the lowest PGS LST decile (online supplemental table 5). For example, at 60 years of age, the cumulative incidence for any CVDs was 44% for those with high PGS LST, while for those with low PGS LST, it was 37% (online supplemental table 7, figure 3). Notably, both groups showed increasing trends, indicating that individuals with low PGS LST were also at risk of incident CVD (figure 3). This higher risk of incident CVD in participants with higher genetic liability replicated in HUNT, except for cerebrovascular disease (HR: 1.08 (95% CI 0.96 to 1.20)) (online supplemental table 6, online supplemental figure 1).
Cumulative incidence curves for high (>90th percentile (red)) and low (<10th percentile (blue)) PGS LST groups in the FinnGen cohort based on Cox proportional hazard models and adjusted for the first 10 principal components, sex and genotyping batch. CVD, cardiovascular disease; PGS, polygenic score for leisure screen time.
Considering other CVDs as competing events attenuated the cumulative incidence estimates for both high and low PGS LST participants, but their difference remained statistically significant for hypertensive diseases and ischaemic heart diseases, although not for cerebrovascular diseases (online supplemental table 8). In further sensitivity analyses with follow-up starting from individual DNA sampling dates (mean follow-up was 6.0 years in FinnGen and 5.0 in HUNT), the associations remained the same or were slightly attenuated in FinnGen and replicated in HUNT except for any CVD and cerebrovascular diseases (online supplemental table 9).
Exploratory analyses showed that the association between PGS LST and incident CVD persisted after adjustments for SES, BMI and smoking status, except for cerebrovascular diseases, where the number of cases was smallest (online supplemental figure 2). Adding the PGSs for CVDs and PGS PA attenuated the observed associations, indicating at least partial pleiotropy between CVD and sedentary behaviour, but not excluding other pathways for the observed effects (online supplemental table 10, online supplemental figure 3). Mediation analysis showed that higher PGS LST was associated with higher self-reported total sitting time (hour/day) in the HUNT cohort (β=0.06, SE=0.02, p=0.016 in a model for hypertensive diseases and β=0.07, SE=0.02, p=0.003 for ischaemic heart diseases) but indicated no potential mediating pathway to incident CVD (online supplemental figure 4). However, higher PGS LST was also associated with lower MET hour/week (β=−0.54, SE=0.09, p<0.001 for hypertensive diseases and β=−0.47, SE=0.09, p<0.001 for ischaemic heart diseases) and higher MET hour/week with lower risk of hypertensive and ischaemic heart diseases (β=−0.01, SE=0.00, p<0.001 and β=−0.01, SE=0.00, p=0.005, respectively). This suggests a potential mediating pathway from genetic liability to sedentary behaviour to incident CVD via insufficient levels of PA.
Discussion
We found that higher genetic liability to LST was associated with more self-reported LST and that the genetic liability to LST also increased the risk of incident CVD, with replicable findings across two independent large populations. The observed effect sizes remained relatively small and attenuated after adjustment, but suggest that the observed adverse health effects may be either directly attributable to genetic effects or mediated by reduced levels of PA.
The underlying role of genetics in sedentary behaviour and morbidity has so far been understudied.8 Evolution has a profound influence on human biology; hence, evolution-based theories provide valuable insights into epidemiology and population health. Evolutionary selection processes may explain why the ‘desire to be sedentary is in our genes’ and also why previously advantageous alleles may currently be disease-causing alleles.9 10 31 In our study, the risk of any CVD increased very consistently by 5% per SD increase in the PGS LST. This magnitude is similar to what has been observed between every hour spent sedentary and risk of fatal and non-fatal CVD (relative risk=1.05, 95% CI=1.02 to 1.07).32 Furthermore, the risk of CVD differed significantly between those in the highest and lowest deciles of PGS LST. Those with high PGS LST had 21% (19% to 24%) higher risk for any CVD than those with low PGS LST, and at the age of 60, 7% higher cumulative CVD incidence. One explanation for these findings may be genetic pleiotropy.
In genetic pleiotropy, the same genes would affect both LST and CVD. This theory is supported by previously published genetic correlations between LST and several CVD-related phenotypes (0.24 for coronary artery disease, 0.32 for overweight, 0.22 for type 2 diabetes, −0.26 for high-density lipoprotein cholesterol and 0.27 for triglycerides), indicating low to moderate common genetic basis between LST and CVDs.15 Previous studies suggest that most robust pleiotropic genes are associated with the function of the nervous system, for example, the DLG4 gene (ENSG00000132535) is associated with synaptic function in the brain, pituitary gland and retina.33 34 Overall, the tissue enrichment analyses suggest that currently known genes associated with LST are expressed mainly in the nervous system, specifically in the brain.15 However, more studies are needed to further elucidate the underlying mechanism between genetic predisposition to LST and the onset of CVDs.
The observed coefficient of determination and effect size between PGS LST and self-reported LST were small, thus potentially raising concerns about how well the score reflects genetic liability to this trait. As there is little prior evidence on how PGSs are associated with sedentary behaviour while some data are available for PA, we discuss our findings aligned to this construct. Our results are consistent with previous studies in which PA PGSs were associated with the corresponding trait and differentiated well the extreme ends of the genetic spectrum, but explain only a small proportion (0.0%–1.4%) of the phenotypic variance.21 35 A plausible explanation for the small variances was assumed to be the heterogeneity in phenotype assessment (ie, PA collected through varying methods). However, in our study, we carefully selected the validation data to mimic the phenotype in the base data. Nevertheless, the fraction of variance explained remained small and may indicate other methodological limitations related to polygenic scoring. One of these is the ‘missing heritability’ dilemma. Missing heritability is the difference between the observed SNP heritability and heritability estimates based on twin studies (ranging from 19% to 56%).36–39 In our case, this is estimated to be at least 12% and potentially caused by the following: inflated family-based estimates,40 relatively small sample sizes of current GWASs,41 inadequate accuracy in phenotyping of the trait of interest and/or the lack of information on less common and rare variants in genotyping arrays used in GWAS studies. More detailed coverage of the variation in the genome by whole genome sequencing is expected to reduce missing heritability.42 43
Policy implications
Our mediation analysis suggests that there may also be an alternative pathway in which sedentary genes contribute to inadequate levels of PA, thereby increasing the risk of incident CVD. Much has been written about the benefits of PA on cardiovascular health,44 with our findings bringing new perspectives to this discussion. Considering that genetics contribute to sedentary behaviour, effective public health strategies should emphasise approaches that nudge people to be active and/or interventions that include a social component and significantly nourish the intrinsic reward system.45 46 Previous work has suggested that systemic problems, like sedentariness, require systemic solutions.46 Whole population approaches to reduce sedentary time and/or increase PA are supported by our findings, where both high and low PGS LST groups showed increasing trends in CVD incidence. Moving from sedentary to active behaviour may confer health benefits, as the human evolutionary background may also explain why physiological functions operate optimally under regular PA.47 We acknowledge that sedentary behaviour may not fully displace PA behaviour,48 and the relationships between genetic liability to sedentary behaviour and observed behaviours throughout the day in different domains require further investigation.
Strengths and limitations
This study has several strengths. We used high-performing and genome-wide methods for genetic scoring and showed the scores to be valid with independent data sets. We used the most comprehensive data available and a robust study design. We also acknowledge that our results should be interpreted with the following limitations in mind. The base data for PGS LST is based on self-reported LST which may be subject to biases from recall, cultural differences and social desirability.49 Future studies including larger sample sizes and device-based measurements of sedentary behaviour are warranted to construct better PGSs and to test the associations with CVDs. Notably, the LST represented screen time activities at the time of the data collection (2011) with limited generalisability to the modern era. The used FinnGen and HUNT may be enriched with CVDs due to participants having had an established contact with healthcare services. The FinnGen did not have data related to lifestyles, and therefore, the exploratory analyses could not be conducted in this cohort. Multiple testing may have increased the risk of Type I error, but it is controlled by the replication design.
Conclusions
The findings of this study confirm our hypothesis that higher genetic liability for sedentary behaviour is associated with a greater risk of developing CVD, although effect sizes with the current data and methods remain small.
Data availability statement
Data are available on reasonable request. Data may be obtained from a third party and are not publicly available. The used FTC subsample data are in the Biobank of the National Institute for Health and Welfare. Data are available for qualified researchers through a standardised application procedure (see the website https://thl.fi/en/web/thl-biobank/for-researchers for details). Access to individual-level genotypes and register data from FinnGen participants can be applied via the Fingenious portal (https://site.fingenious.fi/en/) hosted by the Finnish Biobank Cooperative FinBB (https://finbb.fi/en/). The register data also needs permission from FinData (www.findata.fi). Researchers affiliated with a Norwegian research institution can apply for HUNT data access from the HUNT Research Centre (www.ntnu.edu/hunt) if they have obtained project approval from the Regional Committee for Medical and Health Research Ethics (REC). Researchers not affiliated with a Norwegian research institution should collaborate with and apply through a Norwegian principal investigator. Information on the application and conditions for data access is available online (www.ntnu.edu/hunt/data). The HUNT Databank website provides a detailed overview of the available variables in HUNT (www.ntnu.edu/hunt/databank). Certain data from ancillary HUNT projects may be subject to a time-limited exclusivity provided to the researchers who have financed and conducted the data collection. Biological material is available for analyses, and information on procedures is found on the HUNT Biobank website (www.ntnu.edu/hunt/hunt-biobank). Data from the health registries are not kept by HUNT; instead, linkages between HUNT and registry data must be made for each research project and require that the principal investigator has obtained project-specific approval for such linkage from REC and each registry owner.
Ethics statementsPatient consent for publication
Ethics approval
This study involves human participants and was approved by Finnish Twin Cohort: University of Helsinki (113/E3/01 and 346/E0/05), Helsinki University Central Hospital (136/E3/01, 01/2011, 270/13/03/01/2008, 154/13/03/00/2011, HUS/1799/2017), and the Finnish Institute for Health and Welfare (THL/4743/6.02.04/2021). FinnGen cohort: The Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS) approved the FinnGen study protocol (number HUS/990/2017). The FinnGen study is approved by the THL (approval number THL/2031/6.02.00/2017, amendments THL/1101/5.05.00/2017, THL/341/6.02.00/2018,THL/2222/6.02.00/2018, THL/283/6.02.00/2019 and THL/1721/5.05.00/2019), the Digital and Population Data Service Agency (VRK43431/2017-3, VRK/6909/2018-3 and VRK/4415/2019-3), the Social Insurance Institution (KELA) (KELA 58/522/2017, KELA 131/522/2018, KELA 70/522/2019 and KELA 98/522/2019) and Statistics Finland (TK-53-1041-17). The HUNT study was approved by the Regional Committee for Medical and Health Research Ethics (REC; 2019/29771). Participants gave informed consent to participate in the study before taking part.
Acknowledgments
We thank the participants and all those who contributed to the data collection from the FTC, FinnGen, and HUNT cohorts, without whom this study would not be possible. The detailed list of investigators involved in FinnGen can be found on pages 4–11 of online supplemental file 1.