Despite advancements in preventive measures, (SSIs) remain a significant healthcare challenge due to their high rates of morbidity and mortality. In this study, 104 samples were collected, of which 92.31% showed significant bacterial growth while 7.69% showed no growth, indicating a high incidence of SSIs. This reflects a considerable bacterial load in surgical wounds and highlights a serious concern for postoperative care. Similar finding was reported [27], in Kathmandu, Nepal, although the bacterial growth rate observed here was higher than those reported [28, 29]. These differences may be attributed to variations in hospital hygiene, environmental conditions, and infection control practices. According to [30] reported 77.4% of isolates as MDR, while in our investigation, the proportion was comparable, reflecting the ongoing challenge of antimicrobial resistance in tertiary care settings.
S. aureus was most frequently isolated from patients aged 31–50 years, while P. aeruginosa appeared more often in those aged 10–50 years. Overall, male patients showed higher infection rates than females, a trend also noted [31], who suggested that occupational exposure and lifestyle differences may contribute to this gender disparity.
S. aureus and P. aeruginosa were identified as the two predominant pathogens in surgical site at wound infections. The emergence of antibiotic-resistant strains has made the treatment of these infections increasingly difficult in both hospital and community settings [32]. SSIs are among the most common healthcare-associated infection, with reported prevalence rates ranging from 2.5 to 41.9% depending on the hospital and healthcare systems [33].
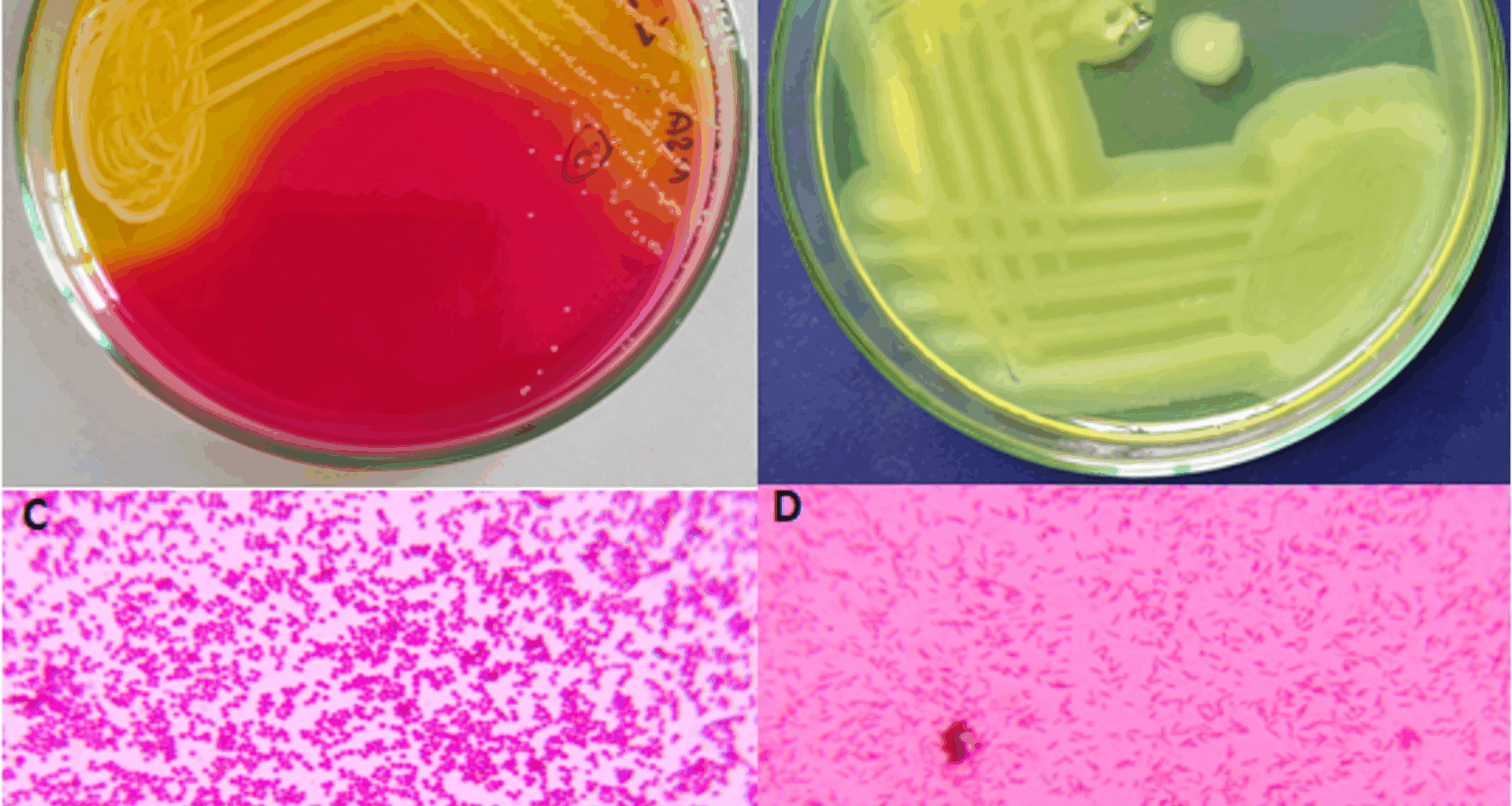
Gram-staining in this study revealed 28.98% Gram-positive and 71.02% were Gram-negative bacteria, which is consistent with findings [34], who reported higher prevalence of Gram-negative bacteria (65.7%) compared to Gram-positive bacteria (34.3%). The rate of infection varies from different country, healthcare settings and even different areas. In our study, S. aureus was 13.10% and P. aeruginosa accounted for 17.40% of infections, findings that were comparable to those reported [35]. Multiple bacterial isolates were observed in 48.08% of the samples, while 44.23% showed single isolates, and 7.69% showed no growth. In contrast [36], reported multiple isolates in only 12.14% of cases. These variations may reflect differences in patient populations, sample sources, and infection control standards. Comparable prevalence of P. aeruginosa (13%) was reported in human pus samples in Egypt, highlighting the consistent global relevance of this opportunistic pathogen in postoperative infections. Variations in prevalence across studies may reflect differences in hospital hygiene, environmental conditions, patient populations, and infection control practices [37].
Biochemical characterization confirmed that all isolates of S. aureus and P. aeruginosa were catalase-positive, citrate-positive, and indole-negative. The catalase positive reaction observed in both organisms aligns with their well-known capacity to produce the catalase enzyme, which essential for protecting against oxidative stress by converting hydrogen peroxide into oxygen and water [18]. Their capacity to use citrate as their only carbon source, further highlights their metabolic flexibility, particularly under nutrient limited conditions, a feature that is especially notable in opportunistic pathogens like P. aeruginosa [21]. Mannitol fermentation, a classical trait of S. aureus. The negative results for the indole test across all isolates reaffirm the established biochemical profiles of these organisms, as neither S. aureus nor P. aeruginosa generally possess the enzyme tryptophanase required for indole production [38].
The antibiotic sensitivity profile of S. aureus isolates revealed complete susceptibility to Linezolid (100%), establishing it as the most effective antibiotic. Moderate susceptibility was observed with Tetracycline, Levofloxacin, Ampicillin/Sulbactam, and Gentamicin, while Ciprofloxacin showed markedly poor activity (11.11%). Among the strains, IIMT-SA17 exhibited the highest sensitivity, as reflected by a large zone of inhibition on the antibiogram. The consistent sensitivity to Linezolid is encouraging and aligns with previous findings [39, 40]. Tetracycline also showed promising activity against the isolates, suggesting that tetracycline resistance genes like tetK and tetM may not be widely prevalent in this region. This is consistent with reports indicating that tetracycline resistance varies significantly depending on local antibiotic usage patterns [41]. Fluoroquinolones such as Levofloxacin showed intermediate sensitivity, while Ciprofloxacin showed the highest resistance rate among all tested antibiotics. This high degree of resistance to Ciprofloxacin most likely reflects mutations in DNA gyrase and topoisomerase IV genes (gyrA, parC), or the operation of efflux pumps, which actively remove the antibiotic from bacterial cells [42]. Ampicillin/Sulbactam showed 50% effectiveness, possibly due to beta-lactamase inhibition by Sulbactam, although resistance suggests the presence of alternative mechanisms such as modified penicillin-binding proteins [43]. Gentamicin was likely due to aminoglycoside-modifying enzymes, commonly reported in nosocomial strains [44]. Multidrug resistance was particularly notable in isolates such as IIMT-SA11, SA06, SA07, and SA04, corroborating earlier reports of S. aureus as a major MDR pathogen in SSIs [45].
Among P. aeruginosa isolates, the highest susceptibility was to Ofloxacin (75%), followed by Piperacillin, Amikacin, Gentamicin, Ampicillin/Sulbactam, and Ciprofloxacin, respectively. Ofloxacin, a fluoroquinolone, appeared to be the most effective, likely due to fewer resistance-conferring mutations in the local population [42, 43]. Piperacillin and Amikacin also showed good activity, consistent with previous reports indicating that β-lactam antibiotics and aminoglycosides remain partially effective against Gram-negative isolates in this region [35].
However, a high level of Ciprofloxacin resistance (87.50%) was observed, which aligns with global reports of P. aeruginosa resistance mediated by gyrA/parC mutations, efflux pump overexpression (e.g., MexAB-OprM), and reduced outer membrane permeability [46, 47]. The most resistant strains (IIMT-PA12, PA13, PA11, and PA07) exhibited resistance to more than three antibiotics, reflecting extensive MDR likely associated with prolonged empirical therapy in clinical settings. These findings are consistent with [36], who reported complete resistance of P. aeruginosa to Erythromycin, Amoxicillin, Colistin, and Cephradine, with retained sensitivity to Imipenem (100%), Apramycin (92.31%), and Amikacin (84.61%).
The high incidence of SSIs and MDR pathogens in this study also points to critical issues in hospital practices and infection control. Lapses in perioperative asepsis, sterilization protocols, and hand hygiene compliance may contribute to elevated infection rates. In addition, widespread empirical use of broad-spectrum antibiotics in tertiary care hospitals likely exerts selective pressure, accelerating resistance development. These findings underscore the urgent need for robust antibiotic stewardship programs, guided by routine antibiogram surveillance and strict adherence to institutional antibiotic policies. Restricting overuse of critical drugs, reinforcing staff training in aseptic techniques, and strengthening postoperative wound care protocols could help mitigate the SSI burden.
Limitations of the current study
There are a few limitations on the current study. It was carried out in a single tertiary care facility with a small sample size, which might limit how broadly the results can be applied. The lack of facilities prevented the molecular characterization of resistance genes, which limited our understanding of the genetic basis of antibiotic resistance. Furthermore, a wider variety of antibiotics and patient-related variables were excluded from the analysis. Notwithstanding these drawbacks, the study offers important new information about the patterns of antibiotic resistance in bacterial pathogens linked to surgical site infections following surgery. However, we are interested in conducting this analysis in the future if appropriate funding support becomes available.