By combining image-based traits with genome-wide association studies (GWAS), the study identified 11 genetic loci—eight of them previously unknown—linked to freeze tolerance. Integrating several of these loci into susceptible wheat germplasm significantly improved freeze resistance, highlighting the potential of AI-driven tools to accelerate the development of more resilient wheat varieties.
Wheat (Triticum aestivum L.) is a cornerstone of global food security, providing essential calories and protein. In major wheat-growing regions, such as China’s Huang-Huai and Yangtze River basins, low-temperature events during winter and early spring frequently damage seedlings. Freeze injury occurs when intracellular ice ruptures membranes and causes tissue death, while late frosts damage spike tissues. Traditional field assessments rely on manual scoring of visible symptoms such as yellowing or tissue collapse. These methods are labor-intensive, subjective, and inconsistent across evaluators, limiting their usefulness for genetic analysis and breeding. As climate variability increases the risk of cold stress, more precise and scalable assessment methods are urgently needed.
A study (DOI: 10.1016/j.plaphe.2025.100061) published in Plant Phenomics on 30 May 2025 by Ni Jiang’s team, Chinese Academy of Sciences, enables accurate, high-throughput assessment of freeze injury in wheat and accelerates the discovery and breeding of cold-tolerant varieties.
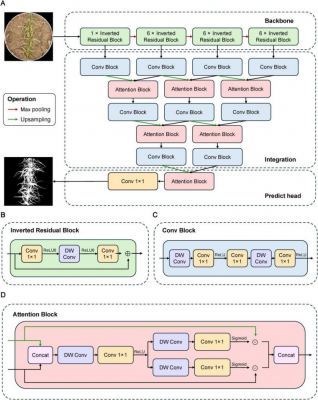
FreezeNet was developed as a lightweight, image-based phenotyping system deployable on smartphones for rapid field evaluation of wheat freeze injury. By incorporating a bidirectional attention mechanism, the model distinguished wheat leaves from complex backgrounds of soil, shadows, and debris with superior accuracy compared to conventional CNN approaches. This architecture generated complementary weight maps, with Attention A optimizing low-level features such as fine textures and color cues, while Attention B refined high-level spatial context and leaf boundaries, thereby reducing misclassification of elements like dead branches or fallen leaves. Benchmarking against UNet, LiteSeg, Lite R-ASPP, and UNeXt revealed that FreezeNet was ~30× smaller and ~42× more efficient than UNet, yet outperformed all models, achieving 94.77% mPA, 81.85% IoU, and 89.95% Dice. From segmented field images, four phenotypic indices—vegetation area (VA), green vegetation area (GVA), yellow vegetation fraction (YVF), and mean hue (mHue)—were extracted and tested across 220 wheat accessions. These image-derived traits showed strong correlations with manual freeze-injury scores (FIS), with YVF emerging as the most reliable indicator (r = 0.80), consistently identifying resistant and susceptible varieties. Importantly, heritability estimates demonstrated that YVF (0.82 ± 0.07) and mHue (0.78 ± 0.08) surpassed FIS (0.64 ± 0.11), making them more powerful for genetic studies. Subsequent genome-wide association studies (GWAS) uncovered 11 quantitative trait loci (QTLs), including eight novel regions, several enriched with C-repeat binding factor (CBF) genes known to regulate cold response. Geographic and temporal analyses further showed resistance alleles clustered in colder provinces and older cultivars. Functional validation in progeny A552, carrying five superior alleles, reduced YVF from 0.88 in the parent line to 0.48, confirming the breeding value of these loci and underscoring FreezeNet’s potential as both a phenotyping and breeding tool for enhancing wheat freeze tolerance.
FreezeNet offers breeders a practical, scalable, and objective tool to assess cold tolerance in wheat and other crops. Its lightweight design enables deployment on resource-limited mobile devices, providing rapid, high-throughput data collection directly in the field. By delivering continuous and quantitative trait data, FreezeNet supports more efficient genetic mapping, allele mining, and molecular breeding. The approach demonstrates how digital agriculture and artificial intelligence can bridge phenotyping bottlenecks in plant science, supporting global efforts to safeguard staple food supplies under climate stress.
###
References
DOI
Original URL
https://doi.org/10.1016/j.plaphe.2025.100061
Funding information
This work was supported by the National Key Research and Development Program of China (2023YFF1000100) and the Biological Breeding-National Science and Technology Major Project (2023ZD04076).
About Plant Phenomics
Plant Phenomics is dedicated to publishing novel research that will advance all aspects of plant phenotyping from the cell to the plant population levels using innovative combinations of sensor systems and data analytics. Plant Phenomics aims also to connect phenomics to other science domains, such as genomics, genetics, physiology, molecular biology, bioinformatics, statistics, mathematics, and computer sciences. Plant Phenomics should thus contribute to advance plant sciences and agriculture/forestry/horticulture by addressing key scientific challenges in the area of plant phenomics.