Risk factors and outcomes of patients with CRPA BSIs
The baseline characteristics of 61 patients with P. aeruginosa BSIs were presented in Table 1. The majority were male (68.8%, n = 42) and elderly (aged > 55 years, 60.6%, n = 37). 70.4% (n = 43) of the patients had at least one hospitalization prior to their first positive culture. Most patients were admitted to ICU (32.7%, n = 20) or hematology ward (37.7%, n = 23), with underlying diseases being mainly malignant tumors (predominantly hematological diseases) in 39.3% (n = 24) of cases, followed by hypertension (19.7%, n = 12) and diabetes (9.8%, n = 6).
Table 1 The characteristics of patients with P. aeruginosa BSIs stratified by carbapenems susceptibility
In the univariate analysis comparing patients with CRPA vs. CSPA BSI, CRPA BSIs was more frequently associated with receipt of mechanical ventilation (p = 0.03), central venous catheter (p = 0.001) and parenteral nutrition (p = 0.003). When entered into a multivariate logistic analysis, central venous catheterization (OR 6.6; 95% CI 1.971–22.104; p = 0.002) was the only independent risk factor for development of CRPA BSI.
During the 30-day follow-up, death occurred in 50.8% (n = 31) of patients with P. aeruginosa BSI. When stratified by carbapenem susceptibility, the 7-day crude mortality rates were 37.2% for CSPA and 38.9% (p = 0.902) for CRPA, while the 30-day rates were 44.2% and 50% (p = 0.678), respectively. Further multivariate Cox regression analysis revealed three factors independently associated with both 7-day and 30-day mortality: prior exposure to carbapenem antibiotics, receipt of mechanical ventilation, and low hemoglobin levels (Table 2 ).
Table 2 Risk factors for 7-day and 30-day mortality in patients with CRPA BSIsAntibiotic resistance spectrum and resistance genes profiles of P. aeruginosa isolates
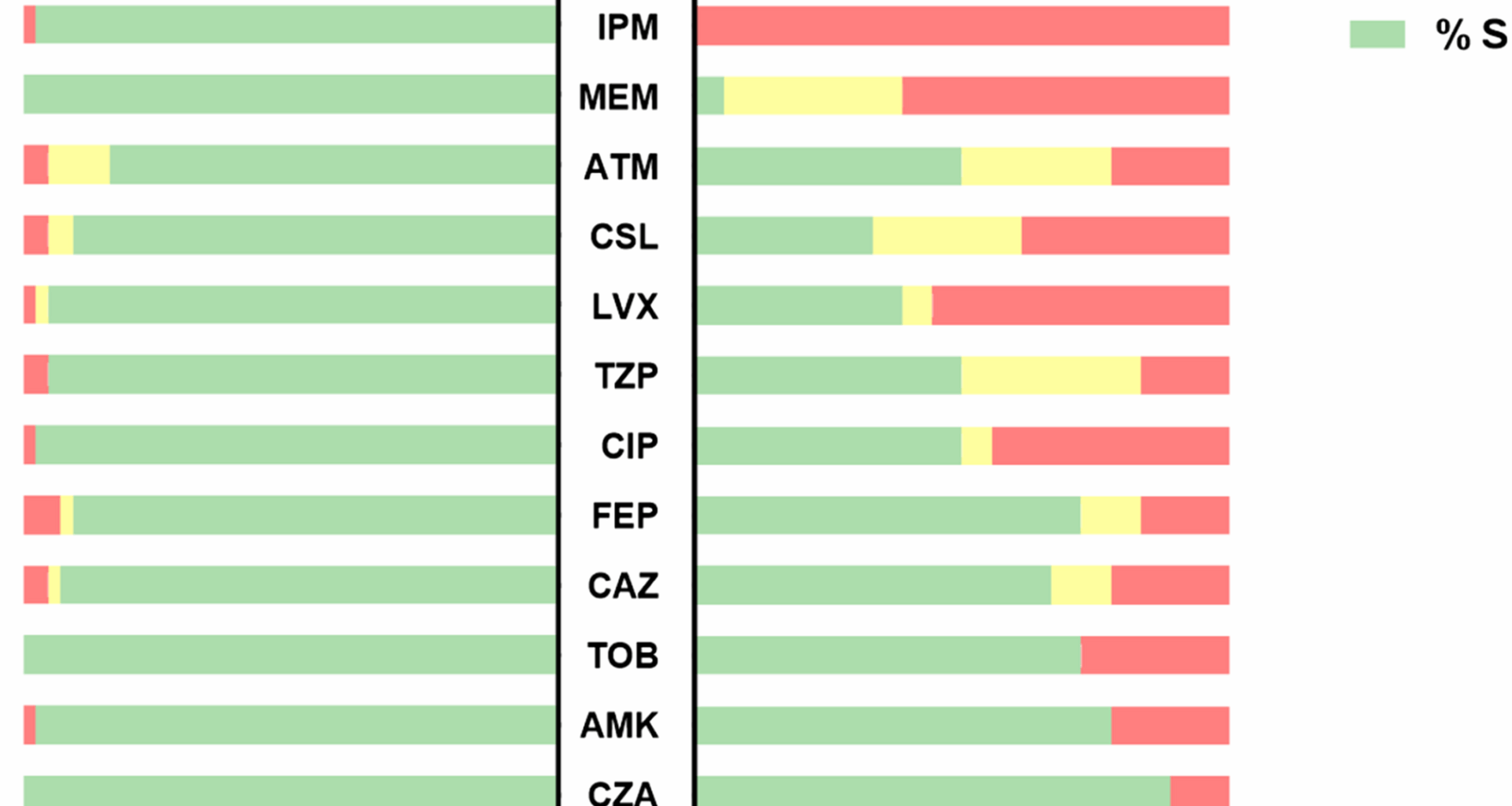
The antimicrobial susceptibility profiles of 61 P. aeruginosa isolates are presented in Fig. 1. The 18 CRPA strains showed significant resistance, with 100% resistance to imipenem and 61.1% resistance to meropenem. Resistance rates to levofloxacin and ciprofloxacin among CRPA strains were 55.6% and 44.4%, respectively. In contrast, lower resistance rates were observed for cefepime (16.7%) and piperacillin/tazobactam (16.7%). The proportion of MDRPA (33.3%, p = 0.002) and DTRPA (16.7%, p = 0.023) strains among CRPA strains were significantly higher compared to CSPA strains.
Antimicrobial susceptibility of 61 P. aeruginosa isolates stratified by carbapenems susceptibility
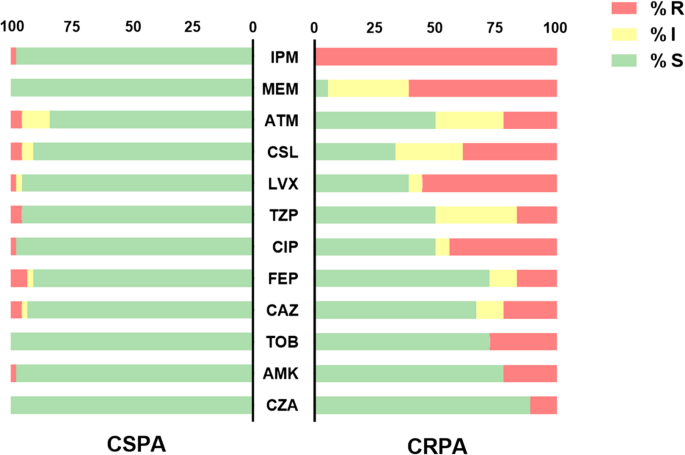
Whole-genome sequencing (WGS) identified a total of 115 AMR genes across 61 P. aeruginosa isolates, with substantial variation in the number of AMR genes carried by individual isolates. Genomic annotation demonstrated that 88.9% (n = 105) of these gene families represented intrinsic chromosomal determinants, including conserved β-lactamase-encoding genes (blaPDC, blaOXA-50, or blaOXA-50-like, etc.); specifically, each strain carried 2 to 4 such conserved β-lactamase genes. Thirteen exogenous AMR gene families were detected, displaying great variability in distribution: only 16.4% (n = 10) of the isolates harbored these exogenous gene familys, with individual carriers containing 0 to 7 genes each. Of particular interest, the presence of these exogenous AMR gene familys was significantly associated with carbapenem resistance (CR) and multidrug resistance (MDR) phenotypes. Among the 61 P. aeruginosa isolates, the most prevalent exogenous AMR genes were AAC(3)-IId (4.9%, 3/61), blaGES-5 (4.9%, 3/61), sul1 (4.9%, 3/61), tet(G) (6.6%, 4/61) and cmlA (8.2%, 5/61). Additionally, five transferable β-lactamase genes included blaKPC-99, blaKPC-2, blaGES-5, blaVEB-1 and blaNDM-5. Most of these β-lactamase genes are located in mobile resistance elements, promoting the spread of resistance traits (fig. 2).
Antibiotic resistance genes, virulence factor determinants, and antimicrobial susceptibility testing results of 61 P. aeruginosa isolates
WGS further revealed divergent carbapenem resistance mechanisms among the 18 CRPA isolates. Six isolates (33.3%) carried β-lactamase genes, including blaKPC-99, blaKPC-2, blaGES-5, blaVEB-1 and blaNDM-5, which exhibited high-level imipenem resistance (MIC ≥ 8 µg/mL). In contrast, the remaining 12 CRPA strains (66.7%) lacked detectable β-lactamase genes, suggesting non-carbapenemase-mediated resistance mechanisms.
Mutational inactivation of OprD is primarily responsible for the resistance to imipenem. Based on sequencing results, carbapenem susceptibility patterns in CRPA strains were stratified according to oprD mutational profiles (Table 3). Almost all CRPA strains exhibited amino acid substitution of oprD. Type 1 included strains with wild-type oprD or silent mutations (preserving amino acid sequences). Remarkably, four CRPA isolates in this group had oprD sequences identical to CSPA strains but remained carbapenem-resistant. Type 2 included strains with significant changes in the oprD amino acid sequence, caused by premature termination codons or frameshift mutations. All isolates in this group demonstrated high-level imipenem resistance (MIC ≥ 8 µg/mL), producing truncated OprD proteins. Among them, clusters 12–14 had oprD point mutations leading to premature translation termination, while clusters15-19 had frameshift mutations in oprD due to nucleotide insertions or deletions. Overall, 44.4% of CRPA strains carried truncated/extended oprD genes (912–1425 bp) instead of the intact 1332-bp form. However, three CRPA strains were excluded from the Type 1 and Type 2 classification because oprD was not detected in their genome sequences.
Table 3 OprD genotypes of 61 P. aeruginosa isolates
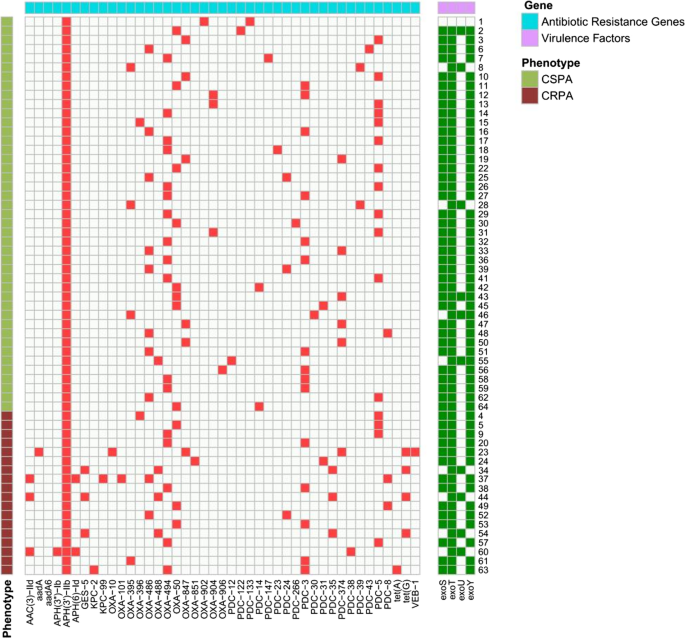
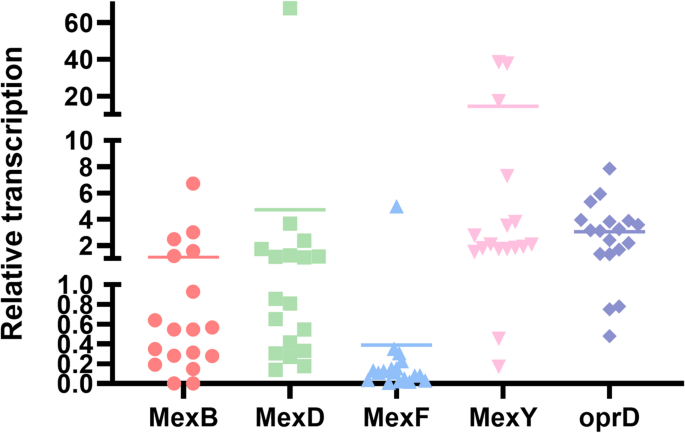
Multidrug efflux pumps are key to carbapenem resistance in P. aeruginosa through extrusion of antimicrobial agents. Using RT-qPCR for gene expression analysis, 61.1% (11/18) of CRPA strains showed significant overexpression (≥ 2-fold change) of efflux pumps (Fig. 3). 50.0% (n = 9) of CRPA strains exhibited increased mexY transcription mRNA levels (2.1- to 38.6-fold). 38.9% (n = 7) of CRPA strains showed downregulated efflux pumps transcription; two of these carried oprD mutations or downregulation, which likely contributes to their carbapenem resistance. However, no known resistance determinants were identified in 22.2% (n = 4) of the CRPA strains.
Relative transcription levels of mex of CRPA strains
Virulence of exoU-positive in CRPA strains
WGS was employed to the intersection of virulence and antimicrobial resistance by genotyping exoU across the 61 P. aeruginosa isolates. The prevalence of exoU showed no significant difference between CRPA and CSPA strains, but a strong association was found when comparing MDRPA with non-MDRPA strains (p = 0.007). Among 17 isolates with oprD mutations, four strains co-harbored exoU+. This proportion was significantly higher compared to that in strains with wild-type oprD phenotypes (p = 0.032). Notably, among these four isolates carrying the oprD mutation and exoU, three also carried blaOXA-488, blaPDC-35, and blaGES-5. These strains exhibited high level carbapenem resistance, with MIC values for imipenem and meropenem both exceeding 8 µg/mL, suggesting a correlation between carbapenem resistance and virulence factor in P. aeruginosa (Table 4).
Table 4 Characteristics of the 10 exoU + P. Aeruginosa strainsEpidemiological analysis of P. aeruginosa isolates
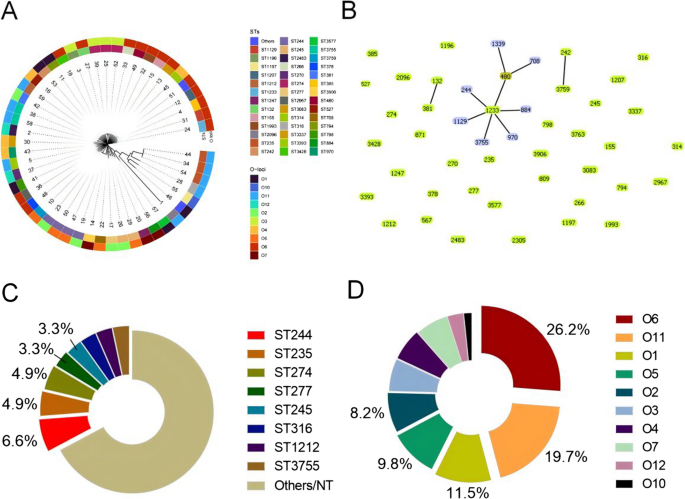
MLST was performed on the 61 P. aeruginosa isolates to analyze their clonal relationships. The results revealed high clonal diversity, with 47 known STs identified among 59 isolates, and 2 new STs among 2 isolates. The most common STs were ST244 (n = 4, 6.5%), ST235 (n = 3, 4.9%), and ST274 (n = 3, 4.9%). Phylogenetic tree of STs revealed 3 clonal complexes (CCs) among the 61 isolates: the largest CC contained 9 STs (ST1233, ST244, ST1129, ST3755, ST970, ST884, ST480, ST1339, and ST708) with 13 strains, while the remaining 2 CCs each included only 2 STs. Of the total 49 STs, 39 were singleton types. CRPA strains displayed sporadic distribution and demonstrated distant genetic relationships (Fig. 4).
Distribution of STs and O-antigen serotypes of 61 P. aeruginosa isolates. (A) Phylogenetic analysis of 61 P. aeruginosa isolates based on MLST and O-antigen serotype; (B) Phylogenetic tree of 61 P. aeruginosa isolates from STs analysis; (C) Pie chart showing sequence type distribution of 61 P. aeruginosa isolates; (D) Pie chart showing O-antigen serotype distribution of 61 P. aeruginosa isolates
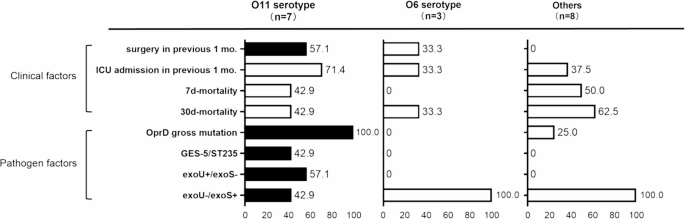
All 61 P. aeruginosa isolates were serotyped using PAst 1.0 based on sequencing data. The most prevalent serotypes were O6 (n = 16, 26.2%) and O11 (n = 12, 19.6%). When comparing resistance groups, the proportion of O11 is significantly higher in CRPA than in CSPA (p = 0.02); this trend was also observed when comparing MDRPA and non-MDRPA (p = 0.004). O6, by contrast, was evenly distributed across resistance phenotypes. O-antigen serotypes also showed distinct associations with T3SS-mediated virulence-resistance profiles (Fig. 5). O11 showed significant correlations with oprD mutations (83.3% vs. 34.5% in non-O11, p = 0.008) and exoU+/exoS-. Non-O11, however, were mainly associated with exoU-/exoS+. Multivariate analysis found no link between O-antigen serotypes and mortality risk, despite these pathobiological differences.
Clinical and pathogen factors in CRPA strains from BSI patients according to O-antigen serotypes. Statistical significance (p ≤ 0.05) is represented by colored bars (black, more prevalent; white, less prevalent)