Quality control and analysis of sequencing data
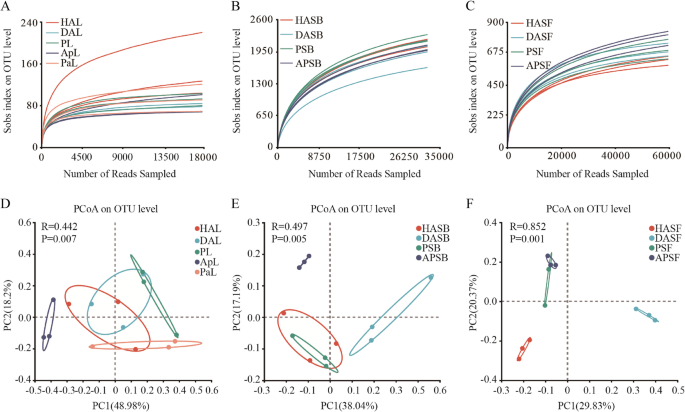
According to the 16 S amplicon sequencing results, a total of 612,606 high-quality bacterial reads were obtained from all leaf samples (Table 1). After removing the low-quality and plant-derived reads, sample leveling was performed by the minimum number of sample sequences, 17,825 reads per sample were retained for subsequent analysis (Figure. 2 A). The remaining reads (267,375) were clustered into 415 endophytic bacterial OTUs at ≥ 97% sequence identity. We used amplicon sequencing of Marker genes to analyze the rhizosphere microbiota of apple and persimmon plants. We obtained 772,169 16 S rRNA gene reads and 841,011 internal transcribed spacer 1 (ITS1) reads. After removing the low-quality and plant-derived reads, sample leveling was performed by the minimum number of sample sequences. A total of 32,316 and 59,637 reads per sample were retained for subsequent analysis (Figure. 2B, C). The remaining reads (387,792 and 715,644) were clustered into 3685 bacterial OTUs and 2187 fungi OTUs at ≥ 97% sequence identity. The rarefaction curves showed that the sequencing work was relatively comprehensive in covering the microbial diversity, as the curves tended to approach saturation, indicating that the selected sequence data adequately reflected the microbial abundance of these samples (Figure. 2 A, C).
Rarefaction curves of the OTU number at 97% similarity for different samples and Bray-Curtis dissimilarity analysis of microbial communities. A, D Endophytic bacteria B, E The bacterial microbiome in the rhizosphere C, F The fungal microbiome in the rhizosphere
On the other hand, the distributions of microbial composition in different apple and persimmon samples were detected on different taxonomy levels, and other diversity indices, including Shannon, Simpson, Ace, and Chao, were applied to estimate the microbial community complexity. All library coverage is above 98.14% (Table S1). Principal coordinate analysis (PCoA) based on Bray-Curtis revealed significant differences (p < 0.007, p < 0.001) in the composition of microbial communities in leaf or soil rhizosphere samples (Figure. 2D, F). The results indicate that apple plants actively recruit distinct microbial communities to colonize the rhizosphere and leaves under healthy, diseased, and intercropped conditions.
Shared and unique microbial communities present during intercropping
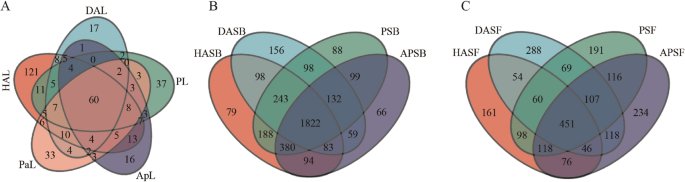
Venn diagram was used to identify the common and unique OTUs among different samples. The OTU abundance of HAL (279) was highest, the secondary was PL (167) and PaL (165), last but one was ApL (136), DAL has least OTU abundance. Five (5) leaves samples shared 60 same OTUs in terms of endophytic bacteria, but harbored 121, 17, 37, 16, and 33 unique OTUs respectively (Figure. 3 A). The OTU abundance of HASB, DASB, PSB, and APSB were 2987, 2691, 3050, and 2735 respectively. Four (4) rhizosphere soils shared 1822 same OTUs in terms of bacteria, but harbored 79, 156, 88, and 66 unique OTUs respectively (Figure. 3B). The OTU abundance of HASF, DASF, PSF, and APSF were 1064, 1193, 1210, and 1266 respectively. In terms of fungi, 4 rhizosphere soils shared 1822 for bacteria same OTUs, but harbored 161, 288, 191, and 234 unique OTUs respectively (Figure. 3 C).
Venn diagrams showing the number of OTUs shared and unique among different samples. A Endophytic bacteria B The bacterial microbiome in rhizosphere C The fungal microbiome in rhizosphere
Comparison of community’s differences at phylum level
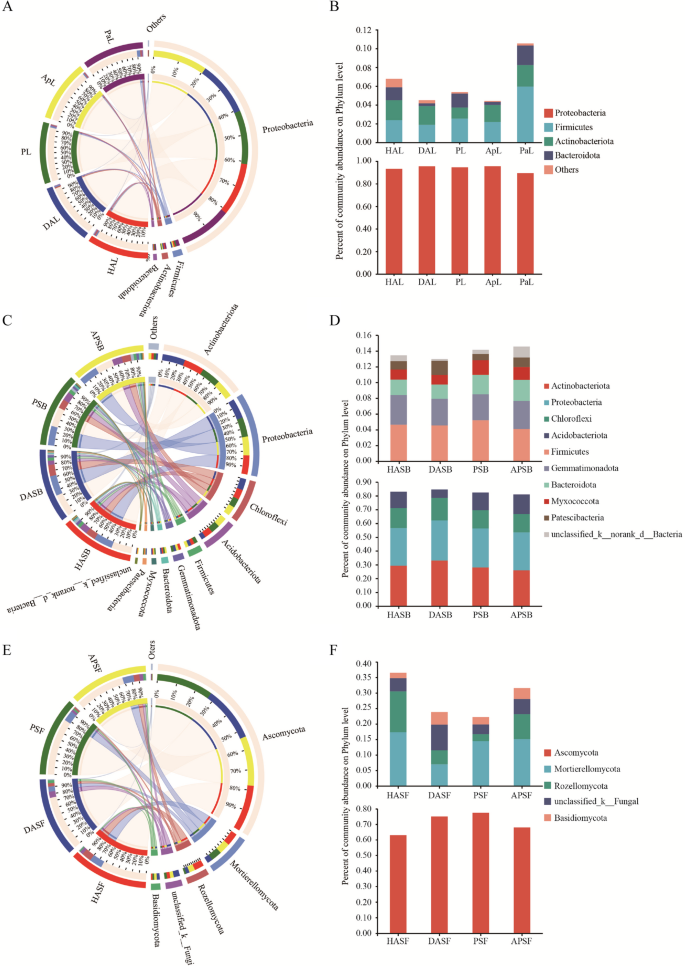
The predominant bacterial components of samples from different treatments were similar (Figure. 4). The phyla showing a relative abundance exceeding 1% were Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes in 5 leaves samples (Figure. 4 A). Proteobacteria was the predominant phylum, representing more than 89% of the total. Firmicutes (1.89%) and Bacteroidetes (0.28%) were the lowest in DAL leaves compared with other leaves samples, while the proportion of Proteobacteria and Actinobacteria in DAL leaves was similar to that of HAL and ApL leaves (Figure. 4B). Actinobacteriota, Proteobacteria, Chloroflexi, Acidobacteriota, Firmicutes, Gemmatimonadota, Bacteroidota, Myxococcota, and Patescibacteria showed a relative abundance higher than 1% in the 4 rhizosphere soils (Figure. 4 C). The proportion of Acidobacteriota and unclassified species in DASB was less than half compared to the other samples. In contrast, the crossed rhizosphere soil in APSB exhibited the highest proportion. In contrast, the three phyla Actinobacteriota, Proteobacteria, and Chloroflexi were slightly higher in DASB than in the remaining samples (Figure. 4D). The fungal phyla with relative abundance higher than 1% were Ascomycota, Mortierellomycota, Rozellomycota, and Basidiomycota (Figure. 4E). Ascomycota was the dominant phylum of all soil samples, accounting for more than 68%. The proportion of Mortierellomycota in DASF was less than half that of the other soil samples, while the proportion of Basidiomycota and unclassified species was higher than in the other soil samples. The proportion of Rozellomycota in HASF (13.18) and APSF(8.03) rhizosphere soil was higher than DASF(4.50) (Figure. 4 F).
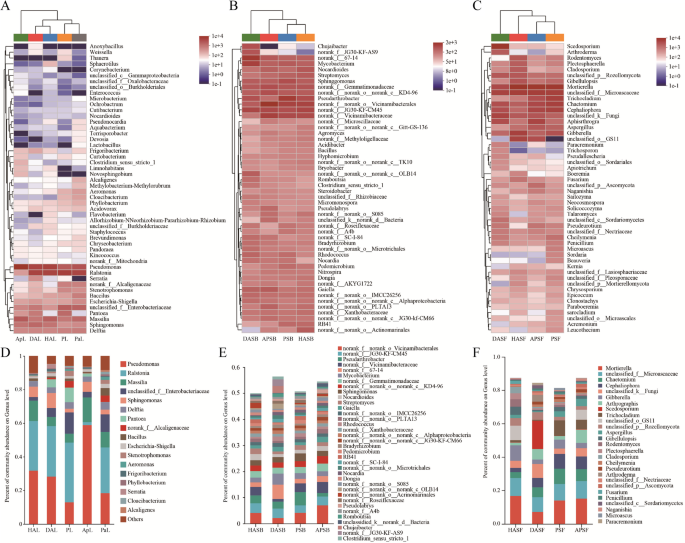
Intercropping regulating the dominant taxa at genus level
The main microbial community for each sample was also analyzed at the genus level (Figure. 5). The results showed that the distribution of each dominant genus among the samples was varied. The heat map and sample cluster tree of leaf endophytic bacteria showed that HAL and DAL clustered into a clade; PL and PaL clustered into a clade, and finally, ApL. This indicates that the diversity cultivation of apple and persimmon trees cross planting can affect the endophytic bacteria of apple leaves. The community structure of leaf endophytic bacteria varied greatly between different groups (Figure. 5 A). At the genus level, the relative abundance of five groups of leaf samples greater than 1% was Pseudomonas, Ralstonia, Massilia, Sphingomonas, Delftia, Pantoea, Bacillus, Stenotrophomonas,
Distribution of microbial community for each sample at phylum level. A, B Leaf. microbial communities C, D Rhizosphere soil bacteria E, F Rhizosphere soil fungi
Aeromonas, Frigoribacterium, Phyllobacterium, Serratia, Cloacibacterim, Alcaligenes, and three unclassified genera (Figure. 5D). The results showed that Pseudomonas had the highest proportion in ApL reaching 58.82% indicating the beneficial interaction among microbes. The proportion of Massilia was also significantly different with HAL (12.18%) and ApL (14.69%) more than twice that of DAL (5.46%). The heatmap and sample clustering tree of rhizosphere soil bacteria showed that PSB and HASB clustered into a clade, next was APSB, and last was DASB. This shows that the soil bacteria diversity can be affected by cross planting with persimmon. The alteration of soil bacteria in the rhizosphere can affect the health of apple trees. Rhizosphere bacterial community structure was quite different between groups (Figure. 5B). At the genus level, the percent of community abundance of four groups of rhizosphere soil samples greater than 1% was Pseudarthrobacter, Mycobacterium, Sphingomonas, Nocardioides, Streptomyces, Gaiella, Rhodococcus, Bradyrhizobium, Pedomicrobium, Nocardia, Dongia, Pseudolabrys, Romboutsia, and Chujaibacter. These relative abundances differed among dominant genera in different rhizosphere soils (Figure. 5E).
At the genus level, the result of the heatmap and the sample cluster tree indicated PSF and APSF clustered into a clade, followed by HASF, and lastly, DASF. This shows that the fungal diversity of apple rhizosphere soil can be affected by cross-planting with persimmon. The alteration of soil fungi in the rhizosphere can affect the health of apple trees, and rhizosphere fungal community structure was quite different between the different groups in partial genus (Figure. 5 C).
At the genus level, the relative abundance of soil fungi in the rhizosphere with more than 1% were Mortierella, Chaetomium, Cephaliophora, Gibberella, Arthrographis, Scedosporium, Trichocladium, Aspergillus, Gibellulopsis, Rodentomyces, Plectosphaerella, Cladosporium, Cheilymenia, Pseudeurotium, Arthroderma, Fusarium, Penicillium, Naganishia, Microascus, and Paracremonium. These dominant genera have different relative abundance in different rhizosphere soil samples. The relative abundance of Mortierella, Gibberella, Aspergillus, Gibellulopsis, and Rodentomyces was significantly lower in DASF than in other rhizosphere soil samples. However, the relative abundance of Scedosporium, Arthroderma, and Paracremonium was higher (Figure. 5 F).
Composition and relative abundance of microorganisms in different samples on the genus level. A Community structure of leaf endophytic bacteria present in different groups B Rhizosphere bacterial community structure in different groups C Rhizosphere fungal community structure in different groups D Relative abundance of leaf endophytic bacteria E Relative abundance of rhizosphere bacteria F Relative abundance of rhizosphere fungi
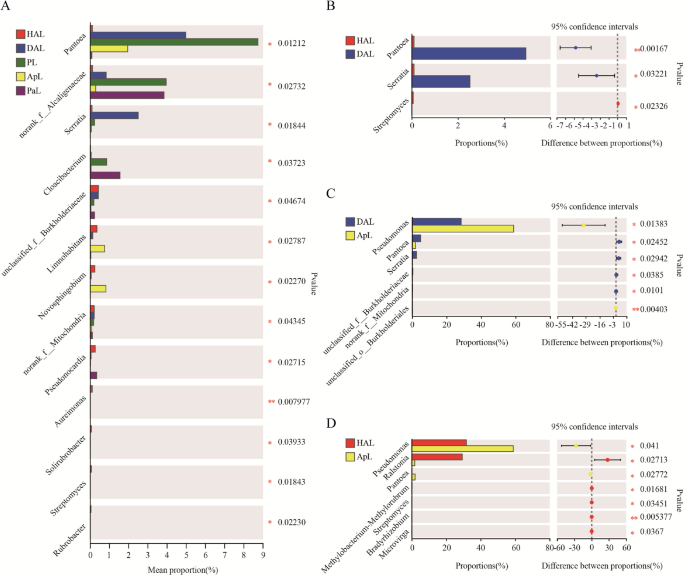
The variation analysis of dominant endophytic bacteria in leaves
Among the five groups of leaf sample, the genus with significant differences (p < 0.05, p < 0.01) were Pantoea, Serratia, Cloacibacterium, Limnohabitans, Novosphingobium, Pseudonocardia, Aureimonas, Solirubrobacter, Streptomyces, and Rubrobacter (Figure. 6 A). Pantoea (p < 0.001) and Serratia (p < 0.03) in DAL were significantly higher than HAL1, while Streptomyces was significantly lower (p < 0.02) than HAL, and did not appear in DAL (Figure. 6B). Pantoea (p < 0.02), Serratia (p < 0.02), and unclassified_f__Burkholderiaceae (p < 0.03) in DAL was significantly higher than ApL, while Pseudomonas (p < 0.01) and unclassified_o__Burkholderiales (p < 0.004) significantly lower than the diversity of ApL (Figure. 6 C). Pseudomonas (p < 0.04) and unclassified_o__Burkholderiales (p < 0.004) in ApL significantly higher than HAL;, Pantoea (p < 0.02), Serratia (p < 0.02), and unclassified_f__Burkholderiaceae (p < 0.03) significantly lower than HAL (Figure. 6D). Notably, all these genera were significantly different (p < 0.05, p < 0.01) than DAL.
Comparison of differences of endophytic bacteria between samples in genus level. A Important group of genera present in all five leaf samples B Different genera in the leaves of healthy and root rot apple tree C Diverse genera in the leaves of root rot apple and combine leaves of apple during intercropping of apple with persimmon D Diverse genera in the leaves of healthy apple and leaves of apple during intercropping of apple with persimmon. Date different symbol mean significant difference between treatments at p < 0.05 and p < 0.01 level
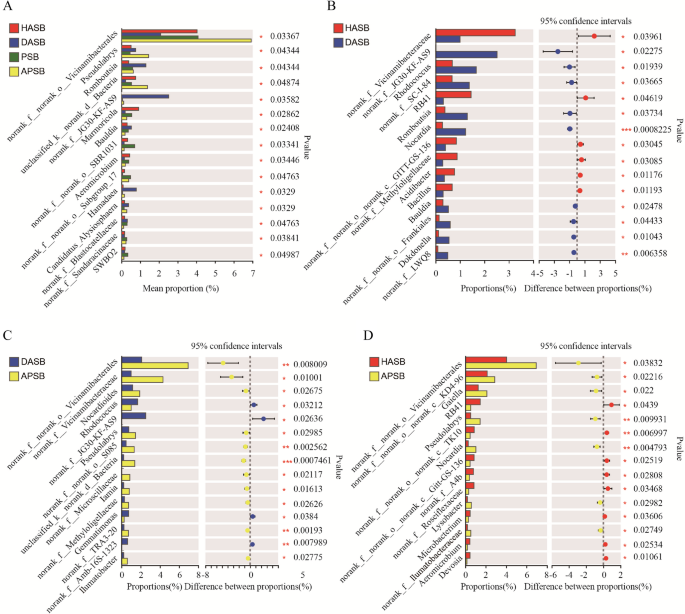
The variation analysis of dominant bacteria in rhizosphere soil
In 4 groups of rhizosphere soil, the top 15 genera with a significant difference (p < 0.05, p < 0.01) were Pseudolabrys, Romboutsia, Marmoricola, Bauldia, Aeromicrobium, Hamadaea, and another genus that were not classified or had no clear taxonomic name (Figure. 7 A). The relative abundance of Acidibacter, Bacillus, and 4 uncategorized genera in HASB was significantly higher (p < 0.05, p < 0.01) than DASB. While Rhodococcus, Nocardia, Bauldia, Dokdonella, Romboutsia, and 4 uncategorized genera significantly (p < 0.05, p < 0.01) lower than DASB (Figure. 7B). Nocardioides, Pseudolabrys, Iamia, Ilumatobacter, and 7 uncategorized genera in APSB was significantly higher (p < 0.05, p < 0.01) than DASB. But Rhodococcus, Gemmatimonas, and 2 uncategorized genus was significantly lower than DASB (Figure. 7 C). Gaiella, Pseudolabrys, Nocardia, Lysobacter, and 3 uncategorized genera in APSB was significantly higher than HASB (Figure. 7D).
Comparison of differences of bacteria between samples in genus level. A Bacterial diversity in rhizosphere in all group B Rhizosphere bacterial diversity in the soil of healthy apple (HASB) and root rot apple (DASB) C Rhizosphere bacterial diversity in the root rot apple and the cross-rhizosphere soil of apple and persimmon D Comparison of rhizosphere diversity in the healthy apple and the cross-rhizosphere soil of apple and persimmon. Different symbols mean significant difference between treatments at p<0.05 and p<0.01 levels
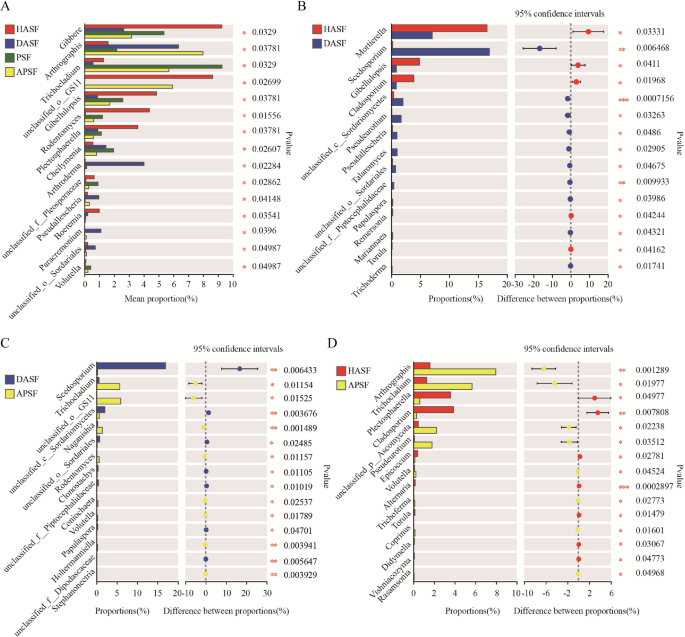
The variation analysis of dominant fungi in rhizosphere soil
In 4 groups of rhizosphere soil, the top 15 genera with a significant difference (p < 0.05, p < 0.01) were Gibberella, Arthrographis, Trichocladium, Gibellulopsis, Rodentomyces, Plectosphaerella, Cheilymenia, Arthroderma, Pseudallescheria, Boeremia, Paracremonium, Volutella, and other genus that was not be classified or have no clear taxonomic name (Figure. 8 A). The relative abundance of Mortierella, Gibellulopsis, Cladosporium, Remersonia, and Torula in HASF was significantly (p < 0.05, p < 0.01) higher than DASF. While Scedosporium, Pseudeurotium, Pseudallescheria, Talaromyces, Papulaspora, Mariannaea, Trichoderma, and 3 uncategorized genus was significantly lower than DASF (Figure. 8B). Trichocladium, Naganishia, Rodentomyces, Coniochaeta, Volutella, Holtermanniella, Stephanonectria, and 1 uncategorized genus in APSF was significantly higher than DASF. Scedosporium, Clonostachys, Papulaspora, and 4 uncategorized genus was significantly lower than DASF (Figure. 8 C). Arthrographis, Trichocladium, Pseudeurotium, Volutella, Trichoderma, Coprinus, Rasamsonia, and 1 uncategorized genus in SF4 was significantly (p < 0.05, p < 0.01) higher than HASF. Plectosphaerella, Cladosporium, Epicoccum, Alternaria, Torula, Didymella, and Vishniacozyma was significantly (p < 0.05, p < 0.01) lower than HASF (Figure. 8D).
Comparison of differences fungal between samples in genus level. A Fungal diversity in rhizosphere in all group B Rhizosphere fungal diversity in the soil of healthy apple (HASF) and root rot apple (DASF) C Rhizosphere fungal diversity in the root rot apple and the cross-rhizosphere soil of apple and persimmon D Comparison of rhizosphere fungal diversity in the healthy apple and the cross-rhizosphere soil of apple and persimmon. Different symbol means significant difference between treatments at p<0.05 and p<0.01