Microbes are a fantastic source of new molecules, many of which are seed compounds for medicinal and agrochemical development. Genomic analysis has revealed that fungi genomes contain many biosynthetic gene clusters, although under normal conditions around 80% of these clusters are silent. Natural product chemists must therefore develop methods to awaken these genes and coax fungi into expressing their secondary metabolites (SMs)—typically by putting them under stress. At present there is no single strategy for achieving this, but genetic engineering, changing the culture conditions, or treating the culture with specific drugs have all shown success.
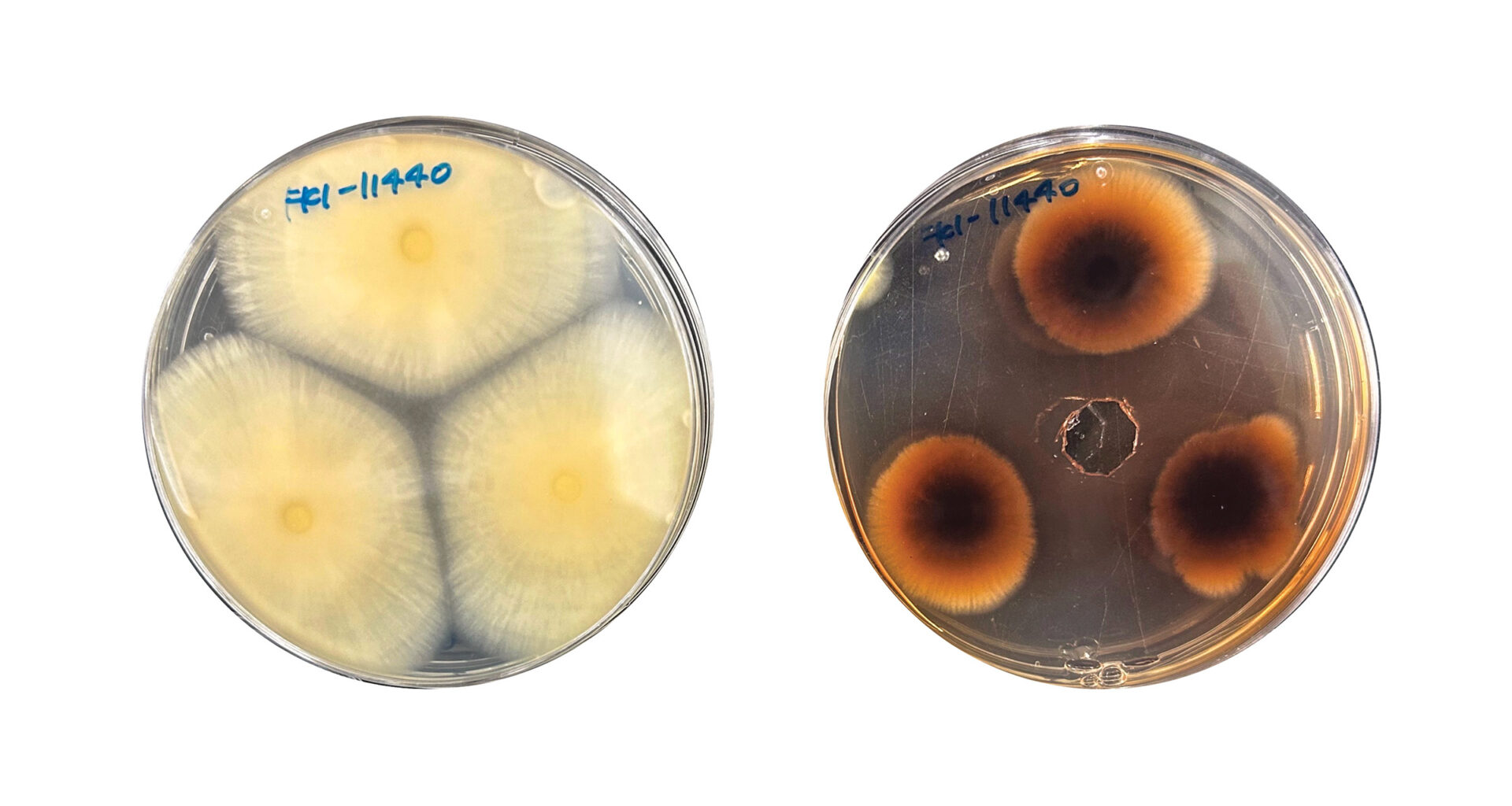
Building on work by Hiroyuki Osada’s group, which reported that the antibiotic hygromycin B induced fungal metabolites in one fungal strain, Masato Iwatsuki, Kenichi Nonaka, and Yoshihiro Watanabe and others at Kitasato University demonstrated that the antibiotic hygromycin B is both an effective and general inducer of fungal metabolites. The antibiotic stimulated compound production in 71% of fungal strains tested and generated three completely novel compounds (J. Nat. Prod. 2025, DOI: 10.1021/acs.jnatprod.5c00770).
The team selected 28 fungal strains from the three classes most commonly isolated from soil—Eurotiomycetes, Sordariomycetes, and Dothideomycetes—and cultured them with and without hygromycin B. The resulting extracts were analyzed by ultra-performance liquid chromatography and mass spectrometry methods. 20 of the 28 strains produced at least 1 compound; the compounds they produced included polyketides, peptides, and terpenes.
“The induced production levels were substantial,” Watanabe says. “In 46% of strains, metabolite yields increased more than threefold, with the maximum reaching a 42.7-fold increase compared to control cultures.”
This initial screen also suggested the presence of several unknown compounds; the team selected two strains to culture on a larger scale to isolate these novel products. After a 14-day culture period, the researchers isolated the terpene hannocateol from Pestalotiopsis sp. and found that it had slight antibiotic activity against the soil bacterium Bacillus spizizenii. Similarly, the strain Pseudoshiraia sp. yielded two terezine analogs, shirazines A and B, which inhibited the growth of both B. spizizenii and a second soil bacterium, Kocuria rhizophila.
The researchers did not detect the three new natural products in samples they later treated with histone deacetylase (HDAC) inhibitors or antifungal drugs but did detect the same products in samples they treated with eukaryotic protein synthesis inhibitors.
“These findings strongly suggest that hygromycin B does not act through chromatin modification like HDAC inhibitors but rather induces SM by interfering with eukaryotic protein synthesis,” Watanabe says.
Neil Kelleher, a proteomics and natural product chemist at Northwestern University, was impressed by the robustness and generality of the approach. He thinks the new inducer provides an attractive alternative to existing induction strategies that will be of particular interest to those working on large-scale screening programs.
“This antibiotic does a pretty good job of teasing out a treasure trove of molecules,” Kelleher says. “Next, you would want to take this to the hundreds or even thousands of strains level.”
The team is now exploring hygromycin B’s applicability to more diverse fungal taxa and hopes to elucidate the precise mechanism of induction. “What’s exciting is that hygromycin B works in a different way from existing methods,” Watanabe says. “The fact that it induced novel compounds that could not be activated by HDAC inhibitors highlights its potential to unlock previously inaccessible SMs, opening a new door to discovering medicines from fungi.”
Chemical & Engineering News
ISSN 0009-2347
Copyright ©
2025 American Chemical Society