World Health O. World malaria report 2024: addressing inequity in the global malaria response. Geneva: World Health Organization; 2024.
Nosten F, White NJ. Artemisinin-based combination treatment of falciparum malaria. Am J Trop Med Hyg. 2007;77:181–92.
WHO. Report on antimalarial drug efficacy, resistance and response: 10 years of surveillance (2010–2019). Geneva: World Health Organization; 2020.
Tripura R, von Seidlein L, Sovannaroth S, Peto TJ, Callery JJ, Sokha M, et al. Antimalarial chemoprophylaxis for forest goers in southeast Asia: an open-label, individually randomised controlled trial. Lancet Infect Dis. 2023;23:81–90.
De Haan F, Amaratunga C, Thi VAC, Orng LH, Vonglokham M, Quang TN, et al. Strategies for deploying triple artemisinin-based combination therapy in the Greater Mekong Subregion. Malar J. 2023;22:261.
Lu F, Culleton R, Zhang M, Ramaprasad A, von Seidlein L, Zhou H, et al. Emergence of indigenous Artemisinin-resistant Plasmodium falciparum in Africa. N Engl J Med. 2017;376:991–3.
Group WKG-PS. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments-a WWARN individual patient data meta-analysis. BMC Med. 2019;17:1
Miotto O, Sekihara M, Tachibana SI, Yamauchi M, Pearson RD, Amato R, et al. Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog. 2020;16:e1009133.
Vanhove M, Schwabl P, Clementson C, Early AM, Laws M, Anthony F, et al. Temporal and spatial dynamics of Plasmodium falciparum clonal lineages in Guyana. PLoS Pathog. 2024;20:e1012013.
Kagoro FM, Barnes KI, Marsh K, Ekapirat N, Mercado CEG, Sinha I, et al. Mapping genetic markers of artemisinin resistance in Plasmodium falciparum malaria in Asia: a systematic review and spatiotemporal analysis. Lancet Microbe. 2022;3:e184–92.
Fola AA, Feleke SM, Mohammed H, Brhane BG, Hennelly CM, Assefa A, et al. Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia. Nat Microbiol. 2023;8:1911–9.
Imwong M, Dhorda M, Myo Tun K, Thu AM, Phyo AP, Proux S, et al. Molecular epidemiology of resistance to antimalarial drugs in the Greater Mekong subregion: an observational study. Lancet Infect Dis. 2020;20:1470–80.
Tumwebaze PK, Conrad MD, Okitwi M, Orena S, Byaruhanga O, Katairo T, et al. Decreased susceptibility of Plasmodium falciparum to both dihydroartemisinin and lumefantrine in northern Uganda. Nat Commun. 2022;13:6353.
Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5.
Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun. 2016;7:11553.
Otienoburu SD, Maïga-Ascofaré O, Schramm B, Jullien V, Jones JJ, Zolia YM, et al. Selection of Plasmodium falciparum pfcrt and pfmdr1 polymorphisms after treatment with artesunate-amodiaquine fixed dose combination or artemether-lumefantrine in Liberia. Malar J. 2016;15:452.
Henriques G, Martinelli A, Rodrigues L, Modrzynska K, Fawcett R, Houston DR, et al. Artemisinin resistance in rodent malaria–mutation in the AP2 adaptor μ-chain suggests involvement of endocytosis and membrane protein trafficking. Malar J. 2013;12:118.
Demas AR, Sharma AI, Wong W, Early AM, Redmond S, Bopp S, et al. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc Natl Acad Sci U S A. 2018;115:12799–804.
Henriques G, Hallett RL, Beshir KB, Gadalla NB, Johnson RE, Burrow R, et al. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis. 2014;210:2001–8.
Henrici RC, van Schalkwyk DA, Sutherland CJ. Modification of pfap2μ and pfubp1 markedly reduces ring-stage susceptibility of Plasmodium falciparum to artemisinin in Vitro. Antimicrob Agents Chemother. 2019;64:1.
Sutherland CJ, Lansdell P, Sanders M, Muwanguzi J, van Schalkwyk DA, Kaur H, et al. pfk13-independent treatment failure in four imported cases of Plasmodium falciparum malaria treated with artemether-lumefantrine in the United Kingdom. Antimicrob Agents Chemother. 2017;61:3.
Sharma AI, Shin SH, Bopp S, Volkman SK, Hartl DL, Wirth DF. Genetic background and PfKelch13 affect artemisinin susceptibility of PfCoronin mutants in Plasmodium falciparum. PLoS Genet. 2020;16:e1009266.
Windle ST, Lane KD, Gadalla NB, Liu A, Mu J, Caleon RL, et al. Evidence for linkage of pfmdr1, pfcrt, and pfk13 polymorphisms to lumefantrine and mefloquine susceptibilities in a Plasmodium falciparum cross. Int J Parasitol Drugs Drug Resist. 2020;14:208–17.
Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47.
Gomez GM, D’Arrigo G, Sanchez CP, Berger F, Wade RC, Lanzer M. Pfcrt mutations conferring piperaquine resistance in falciparum malaria shape the kinetics of quinoline drug binding and transport. PLoS Pathog. 2023;19:e1011436.
Folarin OA, Bustamante C, Gbotosho GO, Sowunmi A, Zalis MG, Oduola AM, et al. In vitro amodiaquine resistance and its association with mutations in pfcrt and pfmdr1 genes of Plasmodium falciparum isolates from Nigeria. Acta Trop. 2011;120:224–30.
Eyase FL, Akala HM, Ingasia L, Cheruiyot A, Omondi A, Okudo C, et al. The role of Pfmdr1 and Pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in western-Kenya P. falciparum samples during 2008–2011. PLoS ONE. 2013;8:e64299.
Verity R, Aydemir O, Brazeau NF, Watson OJ, Hathaway NJ, Mwandagalirwa MK, et al. The impact of antimalarial resistance on the genetic structure of Plasmodium falciparum in the DRC. Nat Commun. 2020;11:2107.
Girgis ST, Adika E, Nenyewodey FE, Senoo Jnr DK, Ngoi JM, Bandoh K, et al. Drug resistance and vaccine target surveillance of Plasmodium falciparum using nanopore sequencing in Ghana. Nat Microbiol. 2023;8:2365–77.
Aranda-Díaz A, Neubauer Vickers E, Murie K, Palmer B, Hathaway N, Gerlovina I, et al. Sensitive and modular amplicon sequencing of Plasmodium falciparum diversity and resistance for research and public health. Sci Rep. 2025;15:10737.
De Cesare M, Mwenda M, Jeffreys AE, Chirwa J, Drakeley C, Schneider K, et al. Flexible and cost-effective genomic surveillance of P. falciparum malaria with targeted nanopore sequencing. Nat Commun. 2024;15:1413.
Wang F, Tian Y, Yang J, Sun FJ, Sun N, Liu BY, et al. Detection of Plasmodium falciparum by using magnetic nanoparticles separation-based quantitative real-time PCR assay. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2014;26:522–5.
Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90.
MalariaGEN, Abdel Hamid M, Abdelraheem M, Acheampong D, Ahouidi A, Ali M, et al. Pf7: an open dataset of Plasmodium falciparum genome variation in 20,000 worldwide samples [version 1; peer review: 3 approved]. 2023;8 22.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Cingolani P, Platts A, le Wang L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92.
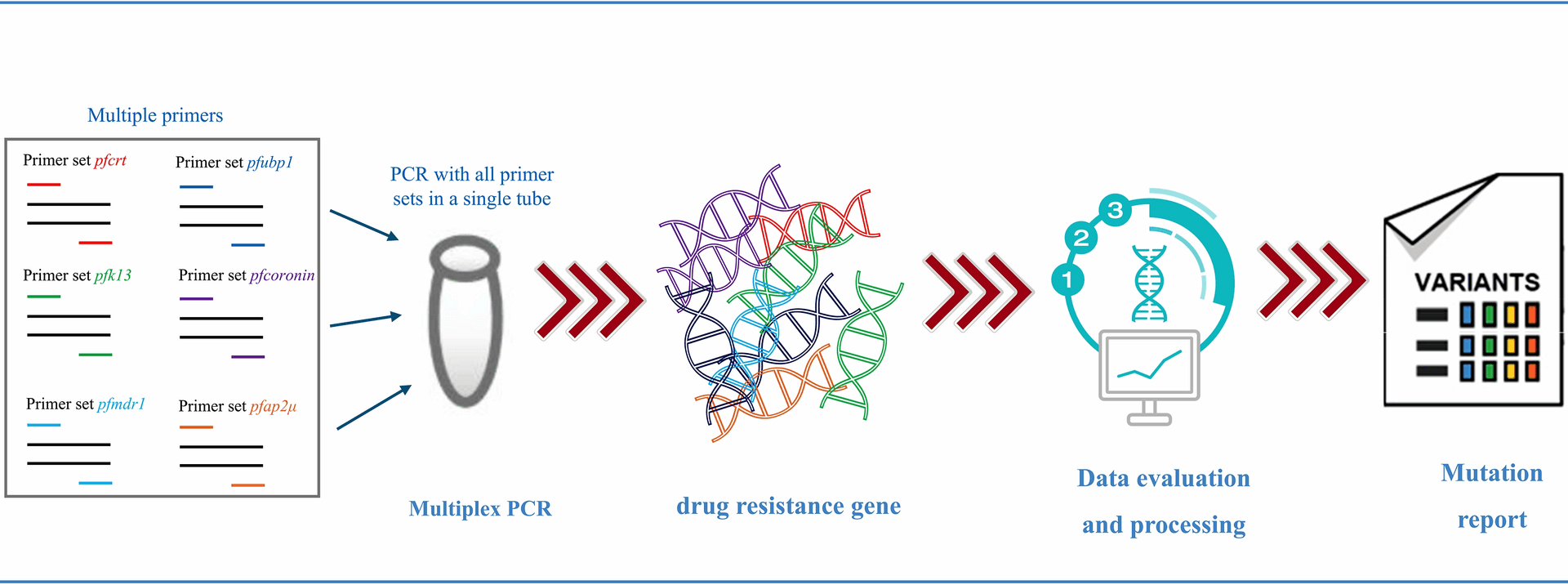
Kattenberg JH, Fernandez-Miñope C, van Dijk NJ, Llacsahuanga Allcca L, Guetens P, Valdivia HO, et al. Malaria molecular surveillance in the peruvian amazon with a novel highly multiplexed plasmodium falciparum AmpliSeq Assay. Microbiol Spectr. 2023;11:e0096022.
Jacob CG, Thuy-Nhien N, Mayxay M, Maude RJ, Quang HH, Hongvanthong B, et al. Genetic surveillance in the Greater Mekong subregion and South Asia to support malaria control and elimination. Elife. 2021;10:1.
Kunasol C, Dondorp AM, Batty EM, Nakhonsri V, Sinjanakhom P, Day NPJ, et al. Comparative analysis of targeted next-generation sequencing for Plasmodium falciparum drug resistance markers. Sci Rep. 2022;12:5563.
Avcı KD, Karakuş M, Kart Yaşar K. Molecular survey of pfmdr-1, pfcrt, and pfk13 gene mutations among patients returning from Plasmodium falciparum endemic areas to Turkey. Malar J. 2024;23:286.
Agaba BB, Travis J, Smith D, Rugera SP, Zalwango MG, Opigo J, et al. Emerging threat of artemisinin partial resistance markers (pfk13 mutations) in Plasmodium falciparum parasite populations in multiple geographical locations in high transmission regions of Uganda. Malar J. 2024;23:330.
Mathieu LC, Singh P, Monteiro WM, Magris M, Cox H, Lazrek Y, et al. Kelch13 mutations in Plasmodium falciparum and risk of spreading in Amazon basin countries. J Antimicrob Chemother. 2021;76:2854–62.
Demirev PA. Dried blood spots: analysis and applications. Anal Chem. 2013;85:779–89.
McDade TW, Williams S, Snodgrass JJ. What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography. 2007;44:899–925.
Adams T, Ennuson NAA, Quashie NB, Futagbi G, Matrevi S, Hagan OCK, et al. Prevalence of Plasmodium falciparum delayed clearance associated polymorphisms in adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1) genes in Ghanaian isolates. Parasit Vectors. 2018;11:175.
Amambua-Ngwa A, Amenga-Etego L, Kamau E, Amato R, Ghansah A, Golassa L, et al. Major subpopulations of Plasmodium falciparum in sub-Saharan Africa. Science. 2019;365:813–6.
Osoti V, Wamae K, Ndwiga L, Gichuki PM, Okoyo C, Kepha S, et al. Detection of low frequency artemisinin resistance mutations, C469Y, P553L and A675V, and fixed antifolate resistance mutations in asymptomatic primary school children in Kenya. BMC Infect Dis. 2025;25:73.
Krishna S, Pulcini S, Moore CM, Teo BH, Staines HM. Pumped up: reflections on PfATP6 as the target for artemisinins. Trends Pharmacol Sci. 2014;35:4–11.
Arya A, Kojom Foko LP, Chaudhry S, Sharma A, Singh V. Artemisinin-based combination therapy (ACT) and drug resistance molecular markers: a systematic review of clinical studies from two malaria endemic regions – India and sub-Saharan Africa. Int J Parasitol Drugs Drug Resist. 2021;15:43–56.