Newswise — Camelina sativa, also known as false-flax, is gaining attention as a biofuel crop due to its potential for aviation fuel production and minimal agricultural input requirements. However, the genetic foundation of this species has been underexplored. Despite its ancient cultivation history, Camelina’s relatively low genetic diversity has hindered breeding programs aimed at improving its yield and stress tolerance. Recent genomic advancements have provided a clearer picture of the crop’s population structure and subgenome composition, offering fresh insights for breeding and biotechnology.
Published (DOI: 10.1093/hr/uhae247) in Horticulture Research, the study by Brock et al. (2024) uncovers new genetic insights into C. sativa, focusing on its subgenome structure and expression patterns. By analyzing a comprehensive diversity panel, the researchers reveal the degree of subgenome dominance and genetic variability across Camelina populations. Their findings, based on the new high-quality Suneson genome, provide crucial information for breeding programs aimed at improving this crop’s performance in biofuel production and agriculture.
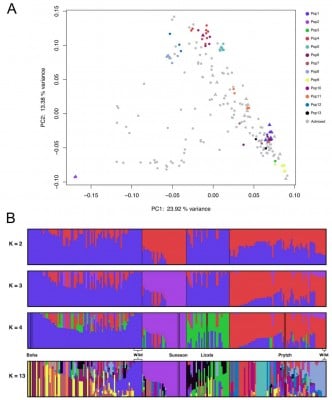
The research utilized advanced genome sequencing to investigate the genetic diversity and subgenome expression of C. sativa. Through the assembly of the Suneson variety genome using PacBio HiFi sequencing, the study identifies three distinct subgenomes and provides an enhanced reference for future genomic studies. The researchers found that despite the overall low genetic diversity, there were 13 distinct subpopulations, including two wild populations. Notably, the SG3 subgenome, which is associated with the C. hispida progenitor, exhibited lower genetic diversity but was more dominant in flower, flower bud, and fruit tissues, critical for the crop’s yield. This study also highlighted the presence of long non-coding RNAs (lncRNAs) in SG3, suggesting its potential role in regulating stress tolerance. These findings challenge previous assumptions about subgenome dominance and provide a more nuanced understanding of Camelina’s genetic architecture. The discovery that subgenome dominance is tissue-dependent has important implications for breeding strategies, particularly in targeting floral and fruit tissues for enhanced yield.
“Understanding the genetic diversity and subgenome expression dynamics in C. sativa is a game-changer for future breeding efforts,” said Dr. Patrick Edger, a co-author of the study. “By identifying the specific subgenome contributions to key traits such as yield and stress tolerance, we can now develop more targeted breeding strategies that enhance crop performance. These insights, combined with the high-quality genome we’ve produced, offer invaluable resources for improving Camelina as a biofuel and agricultural crop.”
The findings from this study have significant implications for Camelina breeding programs aimed at improving yield and stress tolerance. The identification of subgenome-specific expression patterns can guide the development of Camelina varieties with enhanced traits for biofuel production and agricultural applications. The study’s improved genome assembly also opens doors for more efficient genome editing and transgenic research, allowing for precise manipulation of traits. With the growing demand for sustainable biofuels, these advancements position Camelina as a leading candidate in the development of renewable energy sources and provide a model for research in other polyploid crops.
###
References
DOI
Original Source URL
https://doi.org/10.1093/hr/uhae247
Funding information
This work was supported by the Department of Energy Office of Biological and Environmental Research (Grant no. DE-SC0022987 to E.G. and P.P.E.), National Science Foundation (NSF) Postdoctoral Research Fellowship in Biology (PRFB-2109178 to J.R.B.; PRFB-2208944 to K.A.B), National Science Foundation Plant Genome (PGRP-2029959 to P.P.E.), National Science Foundation (IOS-2023310 and NSF DBI-2243562 to A.D.L.N).
About Horticulture Research
Horticulture Research is an open access journal of Nanjing Agricultural University and ranked number one in the Horticulture category of the Journal Citation Reports ™ from Clarivate, 2023. The journal is committed to publishing original research articles, reviews, perspectives, comments, correspondence articles and letters to the editor related to all major horticultural plants and disciplines, including biotechnology, breeding, cellular and molecular biology, evolution, genetics, inter-species interactions, physiology, and the origination and domestication of crops.