By optimizing growth-stage selection and introducing an improved YOLOX-P model, the study achieved precision levels above 95% while identifying novel genetic loci associated with spike number.
Wheat is a cornerstone of the global diet, providing carbohydrates, protein, and dietary fiber for billions of people. Rising populations and evolving diets mean wheat production must increase by at least 60% by 2050. Traditional breeding efforts have boosted yield mainly through kernel size and number, but progress in spike number (SN)—a critical yet complex trait influenced by genetics, planting density, and management—has lagged. Manual counting of spikes in the field is slow, subjective, and impractical for large-scale trials. Advances in computer vision and deep learning have enabled automated detection of plant traits, yet challenges remain in achieving accuracy under real-world field conditions with dense, overlapping spikes. Addressing this gap, the team applied next-generation algorithms to refine spike counting and link phenotypic data to genetic markers for breeding.
A study (DOI: 10.1016/j.plaphe.2025.100051) published in Plant Phenomics on 13 May 2025 by Jindong Liu & Yonggui Xiao’s team, Chinese Academy of Agricultural Sciences (CAAS), offers breeders a powerful tool for high-throughput phenotyping and genetic improvement, paving the way toward greater wheat productivity and global food security.
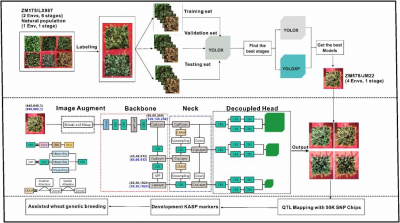
In this study, researchers developed and tested spike number (SN) detection models using images collected across six developmental stages of the Zhongmai 175 × Lunxuan 987 recombinant inbred line (RIL) population, and later validated them in additional populations. Ten models were trained and their performance assessed using four indices—precision, recall, mean average precision (mAP), and F1 score—as well as by comparing labeled spike numbers (ISN), manual counts (MSN), and model-verified spike numbers (VSN). The results showed that precision was consistently high across models (88.19–95.48%), indicating reliable spike identification, but recall varied greatly, from 44.60% to 81.23%. The late grain-filling (LGF) stage consistently delivered the best performance, with the highest recall (81.23%), mAP values (85.69–89.47%), and F1 scores, while early stages were more prone to missed spikes due to leaf obstruction. Comparisons among ISN, MSN, and VSN confirmed that annotation quality strongly influenced accuracy, with the smallest gap between manual and model counts occurring at the LGF stage. Strong correlations (r = 0.84–0.88) were observed between manual and model counts across environments, particularly in Dezhou-trained and Changping-tested models. To further improve detection, an enhanced YOLOX-P algorithm was developed by integrating attention modules and higher-resolution input images. This modification significantly improved performance, increasing mAP by 5.30–5.99% and raising F1 scores by 0.06 compared with the original YOLOX. Among integrated datasets, the CD&DD subset trained with YOLOX-P achieved the highest mAP of 91.81%. Validation in the Zhongmai 578 × Jimai 22 population confirmed the robustness of these models, with correlation coefficients of 0.73–0.82 between automated and manual counts. Genetic analysis further revealed four stable quantitative trait loci (QTLs)—QSN.caas-4A2, QSN.caas-4D, QSN.caas-5B1, and QSN.caas-5B2—linked to spike number. Kompetitive allele-specific PCR markers were developed for two loci, providing valuable tools for marker-assisted breeding. Overall, the YOLOX-P model demonstrated superior capacity to deliver accurate, high-throughput SN phenotyping and enable discovery of novel genetic loci in wheat.
This integration of artificial intelligence with plant genetics represents a step change in wheat breeding. Automated spike counting reduces reliance on labor-intensive field assessments, accelerates phenotyping, and provides objective, scalable data for breeding programs. The development of validated genetic markers offers breeders immediate tools to select varieties with higher spike numbers, directly contributing to yield gains.
###
References
DOI
Original URL
https://doi.org/10.1016/j.plaphe.2025.100051
Funding information
This work was funded by the STI2030-Major Projects (2023ZD04076), the National Natural Science Foundation of China (32372196), the Beijing Joint Research Program for Germplasm Innovation and New Variety Breeding (G20220628002) and the National Natural Science Fund of China (32250410307).
About Plant Phenomics
Plant Phenomics is dedicated to publishing novel research that will advance all aspects of plant phenotyping from the cell to the plant population levels using innovative combinations of sensor systems and data analytics. Plant Phenomics aims also to connect phenomics to other science domains, such as genomics, genetics, physiology, molecular biology, bioinformatics, statistics, mathematics, and computer sciences. Plant Phenomics should thus contribute to advance plant sciences and agriculture/forestry/horticulture by addressing key scientific challenges in the area of plant phenomics.