Melzer R, McCabe PF, Schilling S. Evolution, genetics and biochemistry of plant cannabinoid synthesis: a challenge for biotechnology in the years ahead. Curr Opin Biotechnol. 2022;75: 102684. https://doi.org/10.1016/j.copbio.2022.102684.
Padgitt-Cobb LK, Pitra NJ, Matthews PD, Henning JA, Hendrix DA. An improved assembly of the “Cascade” hop (Humulus lupulus ) genome uncovers signatures of molecular evolution and refines time of divergence estimates for the Cannabaceae family. Hortic Res. 2023;10:uhac281. https://doi.org/10.1093/hr/uhac281.
Schilling S, Dowling CA, Shi J, Hunt D, O’Reilly E, Perry AS, Kinnane O, McCabe PF, Melzer R. The cream of the crop: biology, breeding, and applications of Cannabis sativa. Annu Plant Rev Online. 2021;4:471–528. https://doi.org/10.1002/9781119312994.
Spitzer-Rimon B, Duchin S, Bernstein N, Kamenetsky R. Architecture and florogenesis in female Cannabis sativa plants. Front Plant Sci. 2019;10:350–350. https://doi.org/10.3389/FPLS.2019.00350.
Toth JA, Stack GM, Carlson CH, Smart LB. Identification and mapping of major-effect flowering time loci Autoflower1 and Early1 in Cannabis sativa L. Front Plant Sci. 2022. https://doi.org/10.3389/fpls.2022.991680.
Leckie KM, Sawler J, Kapos P, Mackenzie JO, Giles I, Baynes K, Lo J, Celedon JM, Baute GJ. Loss of daylength sensitivity by splice site mutation in Cannabis. bioRxiv 2023:2023.03.10.532103. https://doi.org/10.1101/2023.03.10.532103.
Garfinkel AR, Wilkerson DG, Chen H, Smart LB, Rojas BM, Getty BA, Michael TP, Crawford S. Genetic mapping of SNP markers and candidate genes associated with day-neutral flowering in Cannabis sativa L. bioRxiv 2023:2023.04.17.537043. https://doi.org/10.1101/2023.04.17.537043.
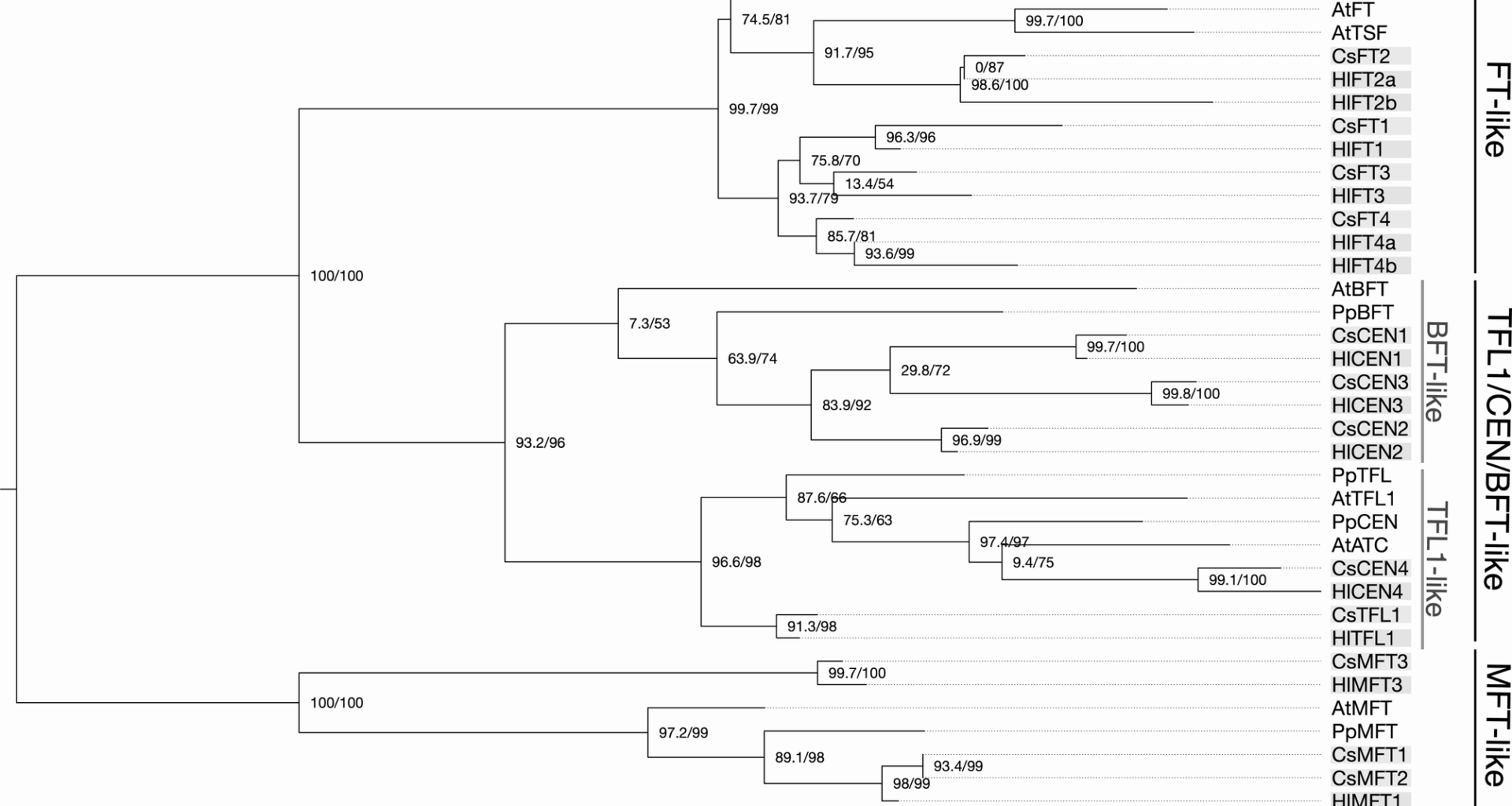
Dowling CA, Shi J, Toth JA, Quade MA, Smart LB, McCabe PF, Schilling S, Melzer R. A flowering locus T ortholog is associated with photoperiod-insensitive flowering in hemp (Cannabis sativa L.). Plant J. 2024;119:383–403. https://doi.org/10.1111/tpj.16769.
Steel L, Welling M, Ristevski N, Johnson K, Gendall A. Comparative genomics of flowering behavior in Cannabis sativa. Front Plant Sci 2023;14:1–22.
Dowling CA, Melzer R, Schilling S. Timing is everything: the genetics of flowering time in Cannabis sativa. Biochemist. 2021;43:34–8. https://doi.org/10.1042/bio_2021_138.
Thomas GG, Schwabe WW. Factors controlling flowering in the hop (Humulus lupulus L.). Ann Bot. 1969;33:781–93. https://doi.org/10.1093/oxfordjournals.aob.a084324.
Prentout D, Stajner N, Cerenak A, Tricou T, Brochier-Armanet C, Jakse J, Käfer J, Marais GAB. Plant genera Cannabis and Humulus share the same pair of well-differentiated sex chromosomes. New Phytol. 2021;231:1599–611. https://doi.org/10.1111/nph.17456.
Fu XG, Liu SY, van Velzen R, Stull GW, Tian Q, Li YX, Folk RA, Guralnick RP, Kates HR, Jin JJ, Li ZH, Soltis DE, Soltis PS, Yi TS. Phylogenomic analysis of the hemp family (Cannabaceae) reveals deep cyto-nuclear discordance and provides new insights into generic relationships. J Syst Evol. 2023;61:806–26. https://doi.org/10.1111/jse.12920.
Heslop-Harrison J. The experimental modification of sex expression in flowering plants. Biol Rev. 1957;32:38–90. https://doi.org/10.1111/j.1469-185X.1957.tb01576.x.
Kobayashi Y, Weigel D. Move on up, it’s time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–84. https://doi.org/10.1101/gad.1589007.
Corbesier L, Coupland G. The quest for florigen: a review of recent progress. J Exp Bot. 2006;57:3395–403. https://doi.org/10.1093/jxb/erl095.
Putterill J, Varkonyi-Gasic E. FT and florigen long-distance flowering control in plants. Curr Opin Plant Biol. 2016;33:77–82. https://doi.org/10.1016/j.pbi.2016.06.008.
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–3. https://doi.org/10.1126/science.1141752.
Golembeski GS, Imaizumi T. Photoperiodic regulation of florigen function in Arabidopsis thaliana. Arabidopsis Book. 2015;13: e0178. https://doi.org/10.1199/tab.0178.
Wickland DP, Hanzawa Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol Plant. 2015;8:983–97. https://doi.org/10.1016/j.molp.2015.01.007.
Gaston A, Potier A, Alonso M, Sabbadini S, Delmas F, Tenreira T, Cochetel N, Labadie M, Prévost P, Folta KM, Mezzetti B, Hernould M, Rothan C, Denoyes B. The FveFT2 florigen/FveTFL1 antiflorigen balance is critical for the control of seasonal flowering in strawberry while FveFT3 modulates axillary meristem fate and yield. New Phytol. 2021;232:372–87. https://doi.org/10.1111/nph.17557.
Xi W, Liu C, Hou X, Yu H. Mother of FT and TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell. 2010;22:1733–48. https://doi.org/10.1105/tpc.109.073072.
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell. 2011;23:3215–29. https://doi.org/10.1105/tpc.111.088492.
Wang Z, Zhou Z, Liu Y, Liu T, Li Q, Ji Y, Li C, Fang C, Wang M, Wu M, Shen Y, Tang T, Ma J, Tian Z. Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell. 2015;27:323–36. https://doi.org/10.1105/tpc.114.135103.
Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH. The role of recently derived FT paralogs in sunflower domestication. Curr Biol. 2010;20:629–35. https://doi.org/10.1016/j.cub.2010.01.059.
Zhang X, Wang C, Pang C, Wei H, Wang H, Song M, Fan S, Yu S. Characterization and functional analysis of PEBP family genes in upland cotton (Gossypium hirsutum L.). PLoS One. 2016;11:e0161080. https://doi.org/10.1371/journal.pone.0161080.
Nelson MN, Książkiewicz M, Rychel S, Besharat N, Taylor CM, Wyrwa K, Jost R, Erskine W, Cowling WA, Berger JD, Batley J, Weller JL, Naganowska B, Wolko B. The loss of vernalization requirement in narrow-leafed lupin is associated with a deletion in the promoter and de-repressed expression of a Flowering Locus T (FT) homologue. New Phytol. 2017;213:220–32. https://doi.org/10.1111/nph.14094.
Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez JM, Lippman ZB. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet. 2017;49:162–8. https://doi.org/10.1038/ng.3733.
Wu F, Sedivy EJ, Price WB, Haider W, Hanzawa Y. Evolutionary trajectories of duplicated FT homologues and their roles in soybean domestication. Plant J. 2017;90:941–53. https://doi.org/10.1111/tpj.13521.
Zhang M, Li P, Yan X, Wang J, Cheng T, Zhang Q. Genome-wide characterization of PEBP family genes in nine Rosaceae tree species and their expression analysis in P. mume. BMC Ecol Evol. 2021;21:32. https://doi.org/10.1186/s12862-021-01762-4.
Venail J, Da Silva Santos PH, Manechini JR, Alves LC, Scarpari M, Falcão T, Romanel E, Brito M, Vicentini R, Pinto L, Jackson SD. Analysis of the PEBP gene family and identification of a novel FLOWERING LOCUS T orthologue in sugarcane. J Exp Bot. 2022;73:2035–49. https://doi.org/10.1093/jxb/erab539.
Li Y, Xiao L, Zhao Z, Zhao H, Du D. Identification, evolution and expression analyses of the whole genome-wide PEBP gene family in Brassica napus L. BMC Genom Data. 2023;24:27. https://doi.org/10.1186/s12863-023-01127-4.
Bennett T, Dixon LE. Asymmetric expansions of FT and TFL1 lineages characterize differential evolution of the EuPEBP family in the major angiosperm lineages. BMC Biol. 2021;19:181. https://doi.org/10.1186/s12915-021-01128-8.
Chardon F, Damerval C. Phylogenomic analysis of the PEBP gene family in cereals. J Mol Evol. 2005;61:579–90. https://doi.org/10.1007/s00239-004-0179-4.
Liu YY, Yang KZ, Wei XX, Wang XQ. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 2016;212:730–44. https://doi.org/10.1111/nph.14066.
Wolabu TW, Zhang F, Niu L, Kalve S, Bhatnagar-Mathur P, Muszynski MG, Tadege M. Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 2016;210:946–59. https://doi.org/10.1111/nph.13834.
Pieper R, Tomé F, Pankin A, Von Korff M. FLOWERING LOCUS T4 delays flowering and decreases floret fertility in barley. J Exp Bot. 2021;72:107–21. https://doi.org/10.1093/jxb/eraa466.
Grassa CJ, Weiblen GD, Wenger JP, Dabney C, Poplawski SG, Timothy Motley S, Michael TP, Schwartz CJ. A new Cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytol. 2021;230:1665–79. https://doi.org/10.1111/nph.17243.
Laverty KU, Stout JM, Sullivan MJ, Shah H, Gill N, Deikus G, Sebra R, Hughes TR, Page JE, Bakel HV, Sciences G, City Y, Technology G, City NY, Centre D, Centre M, Tower W. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Res. 2019;29:146–56. https://doi.org/10.1101/gr.242594.118.Freely.
Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–9. https://doi.org/10.1093/nar/gkl200.
Solovyev V. Statistical approaches in Eukaryotic gene prediction. In: Balding D, Cannings C, Bishop M, editor. Handbook of Statistical genetics. 3rd ed. Wiley-Interscience; 2007. p. 1616. Hoboken, New Jersey, USA.
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–8. https://doi.org/10.1093/nar/gkz991.
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. https://doi.org/10.1093/bioinformatics/btp033.
Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017. https://doi.org/10.1093/bib/bbx108.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. https://doi.org/10.1093/bioinformatics/btp698.
Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–44. https://doi.org/10.1093/nar/gky379.
Verde I, Jenkins J, Dondini L, Micali S, Pagliarani G, Vendramin E, Paris R, Aramini V, Gazza L, Rossini L, Bassi D, Troggio M, Shu S, Grimwood J, Tartarini S, Dettori MT, Schmutz J. The peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics. 2017;18: 225. https://doi.org/10.1186/s12864-017-3606-9.
One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–85. https://doi.org/10.1038/s41586-019-1693-2.
Kong F, Liu B, Xia Z, Sato S, Kim BM, Watanabe S, Yamada T, Tabata S, Kanazawa A, Harada K, Abe J. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 2010;154:1220–31. https://doi.org/10.1104/pp.110.160796.
Wong T, Kalyaanamoorthy S, Meusemann K, Yeates D, Misof B, Jermiin L. AliStat version 1.3 2014.
Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–5. https://doi.org/10.1093/nar/gkw256.
Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. https://doi.org/10.1111/2041-210X.12628.
Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40: e49. https://doi.org/10.1093/nar/gkr1293.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–202. https://doi.org/10.1016/j.molp.2020.06.009.
Shi J, Schilling S, Melzer R. Morphological and genetic analysis of inflorescence and flower development in hemp (Cannabis sativa L.) 2024:2024.01.25.577276. https://doi.org/10.1101/2024.01.25.577276.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. https://doi.org/10.1038/nmeth.3317.
Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–5. https://doi.org/10.1093/bioinformatics/bts356.
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5. https://doi.org/10.1038/nbt.3122.
Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci. 2005;102:7748–53. https://doi.org/10.1073/pnas.0500932102.
Ho WWH, Weigel D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell. 2014;26:552–64. https://doi.org/10.1105/tpc.113.115220.
Nan H, Cao D, Zhang D, Li Y, Lu S, Tang L, Yuan X, Liu B, Kong F. GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS One. 2014. https://doi.org/10.1371/journal.pone.0097669.
Cai Y, Wang L, Chen L, Wu T, Liu L, Sun S, Wu C, Yao W, Jiang B, Yuan S, Han T, Hou W. Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol J. 2020;18:298–309. https://doi.org/10.1111/pbi.13199.
Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006;25:605–14. https://doi.org/10.1038/sj.emboj.7600950.
Page RDM, Charleston MA. From gene to organismal phylogeny: reconciled trees and the gene tree/species tree problem. Mol Phylogenet Evol. 1997;7:231–40. https://doi.org/10.1006/mpev.1996.0390.
Jin J, Yang M, Fritsch PW, Velzen R, Li D, Yi T. Born migrators: Historical biogeography of the cosmopolitan family Cannabaceae. J Syst Evol. 2020;58:461–73. https://doi.org/10.1111/jse.12552.
Lynch RC, Padgitt-Cobb LK, Garfinkel AR, Knaus BJ, Hartwick NT, Allsing N, Aylward A, Bentz PC, Carey SB, Mamerto A, Kitony JK, Colt K, Murray ER, Duong T, Chen HI, Trippe A, Harkess A, Crawford S, Vining K, Michael TP. Domesticated cannabinoid synthases amid a wild mosaic cannabis pangenome. Nature. 2025;643:1001-1010. https://doi.org/10.1038/s41586-025-09065-0.
Jin S, Nasim Z, Susila H, Ahn JH. Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin Cell Dev Biol. 2021;109:20–30. https://doi.org/10.1016/j.semcdb.2020.05.007.
Endo M, Yoshida M, Sasaki Y, Negishi K, Horikawa K, Daimon Y, Kurotani KI, Notaguchi M, Abe M, Araki T. Re-evaluation of florigen transport kinetics with separation of functions by mutations that uncouple flowering initiation and long-distance transport. Plant Cell Physiol. 2018;59:1621–9. https://doi.org/10.1093/pcp/pcy063.
Baumann K, Venail J, Berbel A, Domenech MJ, Money T, Conti L, Hanzawa Y, Madueno F, Bradley D. Changing the spatial pattern of TFL1 expression reveals its key role in the shoot meristem in controlling Arabidopsis flowering architecture. J Exp Bot. 2015;66:4769–80. https://doi.org/10.1093/jxb/erv247.
Prentout D, Razumova O, Rhoné B, Badouin H, Henri H, Feng C, Käfer J, Karlov G, Marais GAB. An efficient RNA-seq-based segregation analysis identifies the sex chromosomes of Cannabis sativa. Genome Biol. 2020. https://doi.org/10.1101/gr.251207.119.
Mohamed R, Wang C-T, Ma C, Shevchenko O, Dye SJ, Puzey JR, Etherington E, Sheng X, Meilan R, Strauss SH, Brunner AM. Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 2010;62:674–88. https://doi.org/10.1111/j.1365-313X.2010.04185.x.
Ryu JY, Lee HJ, Seo PJ, Jung JH, Ahn JH, Park CM. The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Mol Plant. 2014;7:377–87. https://doi.org/10.1093/mp/sst114.
Voogd C, Brian LA, Wang T, Allan AC, Varkonyi-Gasic E. Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit. J Exp Bot. 2017;68:1539–53. https://doi.org/10.1093/jxb/erx044.
Mediavilla V, Jonquera M, Schmid-Slembrouck I, Soldati A. Decimal code for growth stages of hemp (Cannabis sativa L.). J Int Hemp Assoc. 1998;5:68–74.
Schilling S, Melzer R, Dowling CA, Shi J, Muldoon S, McCabe PF. A protocol for rapid generation cycling (speed breeding) of hemp (Cannabis sativa) for research and agriculture. Plant J. 2023;113:437–45. https://doi.org/10.1111/tpj.16051.
Lloyd DG, Webb CJ. Secondary sex characters in plants. Bot Rev. 1977;43:177–216. https://doi.org/10.1007/BF02860717.
Shephard HL, Parker JS, Darby P, Ainsworth CC. Sexual development and sex chromosomes in hop. New Phytol. 2000;148:397–411. https://doi.org/10.1046/j.1469-8137.2000.00771.x.
Liao Z, Zhang T, Lei W, Wang Y, Yu J, Wang Y, Chai K, Wang G, Zhang H, Zhang X. A telomere-to-telomere reference genome of ficus (Ficus hispida) provides new insights into sex-determination. Hortic Res. 2023. https://doi.org/10.1093/hr/uhad257.
Raiyemo DA, Bobadilla LK, Tranel PJ. Genomic profiling of dioecious Amaranthus species provides novel insights into species relatedness and sex genes. BMC Biol. 2023;21:37. https://doi.org/10.1186/s12915-023-01539-9.
Montgomery JS, Giacomini DA, Weigel D, Tranel PJ. Male-specific Y-chromosomal regions in waterhemp (Amaranthus tuberculatus) and Palmer amaranth (Amaranthus palmeri). New Phytol. 2021;229:3522–33. https://doi.org/10.1111/nph.17108.
Aljiboury AA, Friedman J. Mating and fitness consequences of variation in male allocation in a wind-pollinated plant. Evolution. 2022;76:1762–75. https://doi.org/10.1111/evo.14544.
Shi J, Toscani M, Dowling CA, Schilling S, Melzer R. Identification of genes associated with sex expression and sex determination in hemp (Cannabis sativa L.). J Exp Bot. 2025;76:175–90. https://doi.org/10.1093/jxb/erae429.
Liu M, Fernando D, Daniel G, Madsen B, Meyer AS, Ale MT, Thygesen A. Effect of harvest time and field retting duration on the chemical composition, morphology and mechanical properties of hemp fibers. Ind Crops Prod. 2015;69:29–39. https://doi.org/10.1016/j.indcrop.2015.02.010.
Salentijn EMJ, Petit J, Trindade LM. The complex interactions between flowering behavior and fiber quality in hemp. Front Plant Sci. 2019. https://doi.org/10.3389/fpls.2019.00614.
Trubanová N, Pender G, McCabe PF, Melzer R, Schilling S. Exploring phenotypic and genetic variability in hemp (Cannabis sativa) 2023:2023.11.01.565084. https://doi.org/10.1101/2023.11.01.565084.
Zhang M, Anderson SL, Brym ZT, Pearson BJ. Photoperiodic flowering response of essential oil, grain, and fiber hemp (Cannabis sativa L.) cultivars. Front Plant Sci. 2021. https://doi.org/10.3389/fpls.2021.694153.
Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 2009;149:1341–53. https://doi.org/10.1104/pp.108.132134.
Hecht V, Laurie RE, Vander Schoor JK, Ridge S, Knowles CL, Liew LC, Sussmilch FC, Murfet IC, Macknight RC, Weller JL. The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell. 2011;23:147–61. https://doi.org/10.1105/tpc.110.081042.