Prevalence and incidence determined by serological analyses
Of the 360 participants enrolled in the study, 82.2% were female, with median age of 32.5 years. Approximately 32% were physicians, 31% were nurses, 21% were nursing assistants, and 15% were other healthcare professionals. The frontline HCW group had a lower proportion of male, were younger, and had a greater number of nurses than physicians. Approximately a half of frontline HCWs worked in the Department of Medicine, mainly in inpatient wards (IPD) (Table 1). Most participants completed the last blood drawn, which were 217 (90%) for frontline HCW and 103 (86%) for non-frontline HCW group. Rapid test by ECLIA for qualitative anti-SARS-CoV-2 NP antibodies showed positive for one non-frontline HCW at the day of enrollment and remained positive across all four visits. This individual underwent RT-PCR test for SARS-CoV-2 upon the initial positive rapid antibody test and yielded a negative result. The rapid test results for the remaining 359 volunteers were consistently negative throughout the study, up to the final visit (Visit 4).
Table 1 Demographic data of frontline and non-frontline healthcare worker
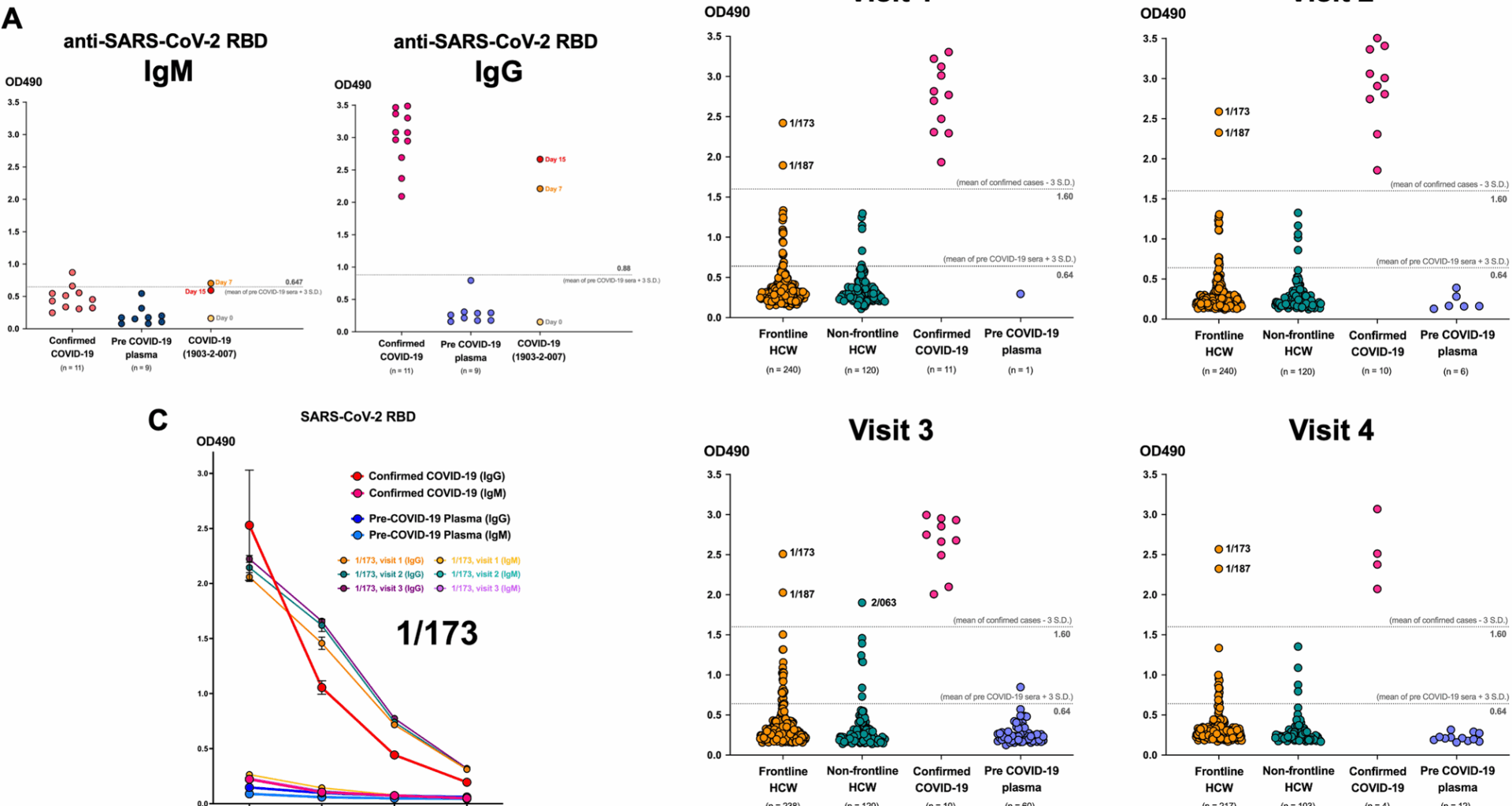
At the beginning of COVID-19 pandemic when approved commercial high-throughput serological methods were not widely available, an in-house conventional ELISA assay for antibodies against RBD of SARS-CoV-2 S protein was set up for serological surveillance and research. The reliability of the assay was verified with the plasma samples of confirmed COVID-19 cases for both IgM for acute infection and IgG for longer follow up. We found that IgG showed higher sensitivity and specificity and correlated well with the antibody levels in confirmed cases when using 3 standard deviation (SD) of the OD490 of pre-COVID-19 sera as the cut-off level (Fig. 1A). Therefore, we performed the ELISA for anti-SARS-CoV-2 RBD IgG was performed to determine the seroprevalence of SARS-CoV-2 infection of all participants across all four visits. The negative cut-off was OD490 = 0.64, which was 3SD above the mean OD490 of pre-COVID-19 sera and the positive cut-off was OD490 = 1.60, which was 3SD below the mean OD490 of confirmed COVID-19 sera. This were based on the experiment of visit 3, when we used the highest number of pre-COVID-19 sera (n = 60). We found that two frontline HCWs (1/173 and 1/187) showed significantly elevated levels of anti-SARS-CoV-2 RBD comparable to the confirmed cases since visit 1. Their levels remained high across all four visits, persisting up to 48 weeks (Fig. 1B). This indicates a seroprevalence of 0.55% (2/360) among all HCWs and 0.83% (2/240) among frontline HCWs. Additionally, one non-frontline HCW (2/063) exhibited a significant increase in antibody levels, similar to infected patients at visit 3. This could assume a SARS-CoV-2 infection incidence, determined by serological test, of 0.28% (1/360) among all HCWs and 0.83% (1/120) among non-frontline HCWs. However, this individual did not attend Visit 4. Importantly, none of these three individuals had prior history of COVID-19 or positive RT-PCR for SARS-CoV-2. Interestingly, the one who tested positive by rapid ECLIA test throughout the study consistently showed negative anti-SARS-CoV-2 RBD IgG ELISA results for all 48 weeks. Moreover, a 3-fold serial dilution of participant 1/173’s plasma showed consistently high level of anti-SARS-CoV-2 RBD IgG comparable to that of the COVID-19 patient at all three visits (Fig. 1C), suggesting that this could be pre-existing antibodies that cross-react with the RBD of SARS-CoV-2.
Two frontline heath care workers (HCWs) with the plasma level of anti-SARS-CoV-2 IgG comparable to confirmed COVID-19 patients at all four visits. (A) Dot plots showing the validation of the ELISA assays for anti-SARS-CoV-2 IgM and IgG at 1:100 dilution using plasma of confirmed COVID-19 patients and plasma collected before COVID-19 pandemic. (B) Dot plots showing the level of anti-SARS-CoV-2 RBD IgG at the 1:100 dilution of plasma determined by ELISA. The number by the dots representing the assigned number for individual study subjects. ELISAs of all samples in each visit were performed at the same time. (C) Three-fold serial dilution for anti-SARS-CoV-2 RBD IgM and IgG of plasma from a frontline HCW (subject 1/173) compared to the plasma of a confirmed COVID-19 patient and a person collected before COVID-19 pandemic
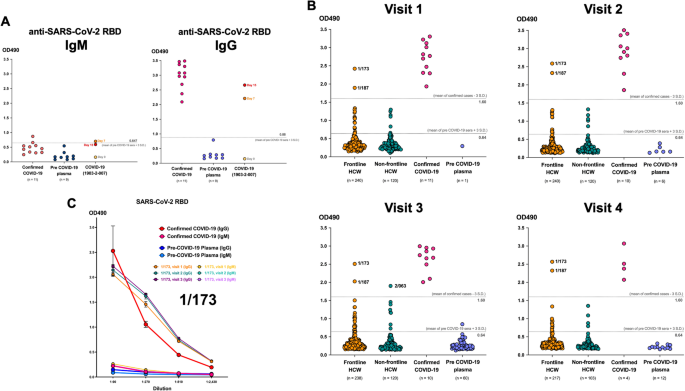
We found that 13–21 frontline HCWs and 4–7 non-frontline HCWs had antibody levels categorized as “borderline” with the OD490 above the negative cut-off but below the positive cut-off (Fig. 1B). To investigate whether these positive and borderline levels of anti-SARS-CoV-2 RBD IgG could reflect the actual level of antibodies and could suggest the SARS-CoV-2 infection, we compared the OD490 to the approved quantitative CMIA for anti-SARS-CoV-2 RBD IgG (Fig. 2A). Of all 175 frontline and 65 non-frontline HCWs, there was a significant correlation only in the frontline HCW group (r = 0.2676, p = 0.0003) and the correlation was still significant when we considered only the ones with positive and borderline groups (r = 0.8588, p = 0.0003). Further microneutralization assay was performed to confirm the SARS-CoV-2-specific antibodies in samples with positive and borderline anti-SARS-CoV-2 RBD IgG and all of them showed negative results at one or all three visits (Fig. 2B). In addition, we further performed a 3-way comparison of serological assays used in our study for IgG antibodies against SARS-CoV-2 (Table S1). The negative percent agreement (NPA) between these assays were very high (> 90%), but the positive percent agreement (PPA) was 0% for all assays.
Correlation between positive and borderline level of anti-SARS-CoV-2 RBD IgG in healthcare workers detected by ELISA and CMIA or SARS-CoV-2 microneutralization assay. (A) Graphs demonstrating the correlation between the level of anti-SARS-CoV-2 RBD by ELISA and CMIA at visit 4. Upper right graph showing only the samples considered positive and borderline by ELISA. (Pearson r with p value were indicated in the graphs). (B) Table demonstrating the neutralization titer (NT50) of antibodies in samples against SARS-CoV-2. The titer lower than 10 is considered negative
Cross-reactivity of pre-existing antibodies against common cold coronaviruses to the spike protein of newly emerged SARS-CoV-2
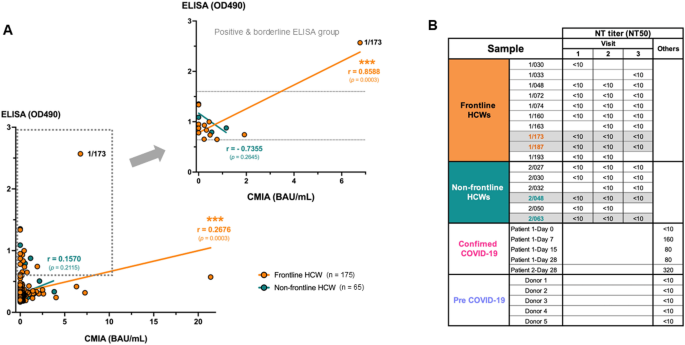
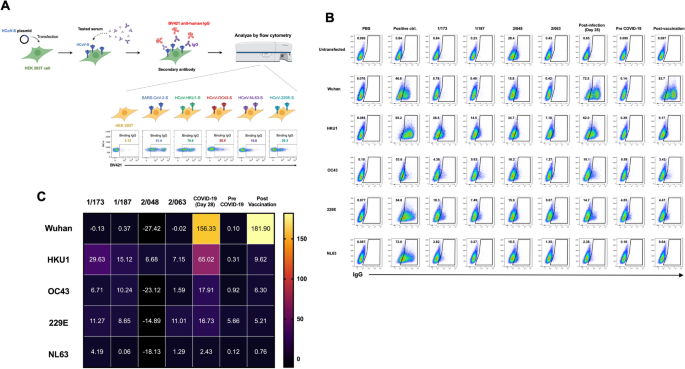
To further investigate whether pre-existing IgG antibodies against other HCoVs could contribute to the positive level of anti-SARS-CoV-2 RBD by conventional ELISA, flow cytometry-based assay for cross-reactivity of antibodies against other HCoV S proteins was performed (Fig. 3A) [10]. This assay allows the detection of antibodies that can bind to the S protein in more physiological forms that are expressed on the phospholipid bilayer membrane of the viral envelope. The plasma samples of two subjects from the frontline HCW group (1/173 and 1/187) that showed high OD490 at all four visits comparable to those in confirmed COVID-19 cases, together with another two subjects from non-frontline HCW group with high borderline OD490 (2/048 and 2/063) as showed in Fig. 2B were tested. Figure 3B demonstrates the percentage of cells expressing S proteins of different HCoVs that the IgG in the plasma samples could bind to. Gating was based on the untransfected cells in the absence of any antibodies. The plasma of subject 2/048 showed high background to the untransfected cells with 28.4% of IgG-bound cells. The expression of each S protein on transfected cells was evaluated by using monoclonal human anti-SARS-CoV2 S1 (CR3022) and polyclonal rabbit antibodies specific for the S1 of each strain of HCoVs and showed as positive control with the satisfied expression level ranging between 53.6% and 95.2% for the S protein of HCoVs.
The cross-reactivity to other human coronaviruses in HCWs with high level of anti-SARS-CoV-2 RBD by ELISA. (A) Diagram of flow cytometry-based assay for cross-reactivity of antibodies in the plasma of study subjects to the spike proteins of other circulating HCoVs. (B) Pseudocolor plots showing the percentage of spike protein-expressing cells that were bound by antibodies in the plasma of study subjects compared with plasma of confirmed COVID-19 patients at day 28 after diagnosis, COVID-19 vaccinee and plasma collected before COVID-19 pandemic. (C) Heat map demonstrating the percentage of human antibody-bound cells compared with that of positive control using rabbit antibodies against spike protein of each human coronavirus. The percentage of antibody-bound untransfected cells were used for background subtraction
The accuracy of this assay in detection of the binding of SARS-CoV-2-specific IgG in the plasma to the S proteins was evaluated by using plasma from a COVID-19 patient at 28 days after diagnosis (CMIA 206.72 BAU/mL) and a volunteer who received a booster COVID-19 mRNA vaccine for 18 days (CMIA 1102.83 BAU/mL). These samples contain IgG that could bind with Wuhan S protein-expressing cells at 72.5% and 83.7%, respectively, compared with 0.65% and 0.1% of the untransfected cells. These were higher than that of the monoclonal antibody positive control (46.8%), which is not surprising as the antibodies induced by infection or vaccination are polyclonal that could bind to more different epitopes of the S proteins on the cells. In addition, plasma from a volunteer collected in July 2015, representing the pre-COVID-19 plasma showed only 0.14% of positive cells. Altogether these results suggest the high accuracy level of this assay.
Therefore, the cross-reactivity of antibodies in the samples was determined by the percentage of IgG-bound cells that expressed different HCoVs compared to those of positive controls (Fig. 3C). We found that all selected samples showed no level of IgG that could bind to the SARS-CoV-2 S protein. All two samples from the frontline HCW group had the highest percentage of IgG-bound HCoV-HKU1 S protein expressing cells at 29.63% for the subject 1/173 and 15.12% for the subject 1/187. Moreover, plasma of the subject 1/173 contained IgG that could also bind to the S protein of HCoV-229E at 11.27%, whereas plasma of the subject 1/187 contained IgG that could bind to the S protein of HCoV-OC43 at 10.24%. Subject 2/063 from non-frontline HCW group had IgG that bound to HCoV-229E S protein the most with 11% positive cells. It is worth noting that natural infection induced the IgG antibodies with higher cross-reactivity to S proteins of other HCoVs (65% for HCoV-HKU1, 18% for HCoV-OC43 and 17% for HCoV-229E) compared with vaccination even with the higher level of binding IgG to the SARS-CoV-2 RBD detected by CMIA. The cross-reactivity to the HCoV-NL63 S protein was low in all tested samples. Therefore, the high level of anti-SARS-CoV-2 RBD by conventional ELISA in the HCWs in this study is highly possible due to the pre-existing antibodies to other HCoVs, especially the HCoV-HKU1 that is in the same betacoronavirus genus as the SARS-CoV-2.