The incidence of SUA was identified to be approximately 2.45% in this study, which is similar to previous findings [5, 17]. SUA has been widely recognized as a prenatal marker associated with an elevated risk of both congenital anomalies and chromosomal abnormalities [6, 7, 18]. In addition, a previous study conducted by Murphy-Kaulbeck et al. [19] indicated that fetuses and neonates with SUA had a 6.77 times greater risk of congenital anomalies and as high as 15.35 times greater risk of chromosomal abnormalities. Despite these associations, limited studies have explored the genetic underpinnings of SUA, and its precise etiology and pathogenesis remain poorly defined. In the present study, a total of 123 fetuses identified with SUA underwent detailed prenatal genetic evaluation, using both conventional karyotyping and CMA, to clarify genotype–phenotype associations and deepen understanding of the genetic underpinnings of SUA.
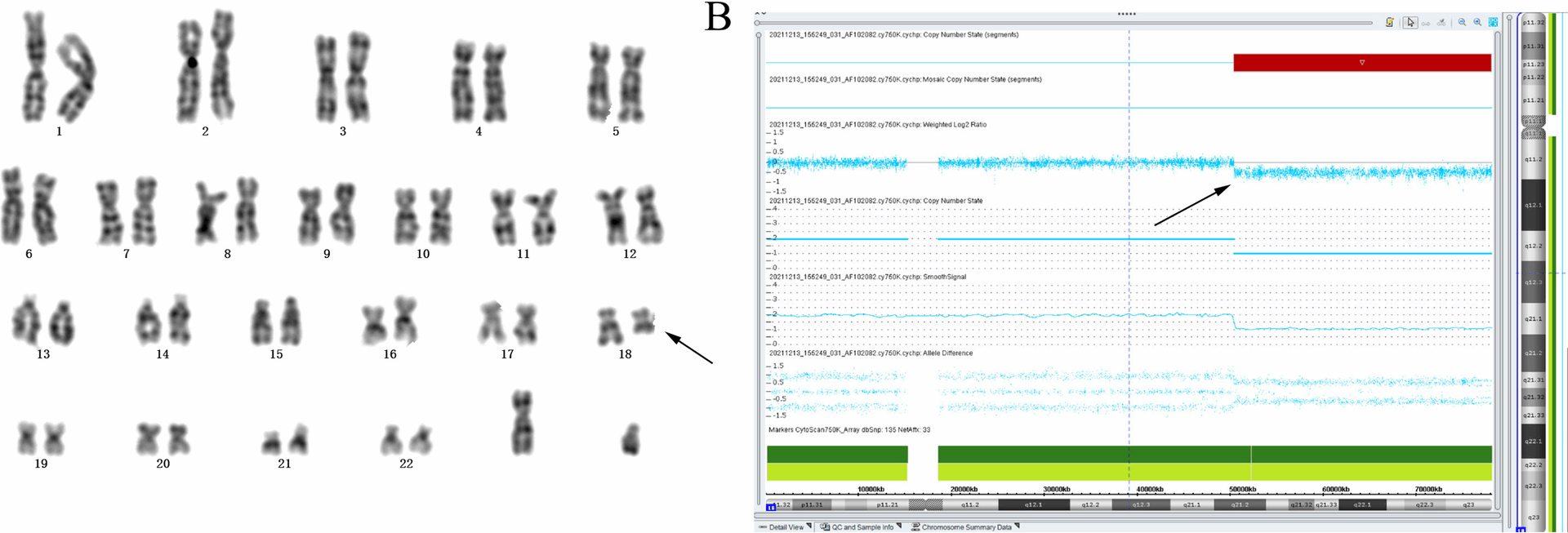
Multiple studies have pinpointed trisomy 18 as the most common aneuploidy in fetuses with SUA [20, 21]. In the present cohort, karyotype analysis uncovered four aneuploidies: two trisomy 18 cases, one monosomy X (45,X), and one 47,XYY. Both trisomy 18 fetuses exhibited SUA alongside additional soft-marker anomalies, while the sex chromosome aneuploidies presented with isolated SUA, corresponding to an 8.00% aneuploidy rate within the isolated SUA subgroup. The lack of extensive population-based data, however, hampers precise quantification of the link between isolated SUA and chromosomal aneuploidies. Previous case reports have also suggested a potential link between structural chromosomal abnormalities and SUA. For instance, a study identified a fetus with a deletion at 18q21.3-qter and a terminal duplication of 1p36.32-p36.33, associated with multiple congenital anomalies including SUA [22]. Similarly, Cho et al. [23] described a fetus with partial trisomy (18pter→q22) and partial monosomy (18q22→qter), also presenting with multiple anomalies including SUA. In the present study, one case with a de novo deletion was identified in the long arm of chromosome 18 [46,XN, del(18)(q21)], associated with SUA and other soft marker anomalies. This finding further supports a potential correlation between long arm abnormalities of chromosome 18 and the presence of SUA. In addition, a chromosomal structural abnormality was identified in a fetus with isolated SUA, characterized as 46,XN, rec(4)dup(4p)inv(4)(p15.1q34.3)dpat. Notably, a previous study reported a 17.8 Mb deletion in the 4q34.1–q35.2 region in a fetus presenting with SUA in combination with a ventricular septal defect and a small midline brain cyst. This suggests a possible association between microdeletions involving the distal region of chromosome 4q and the occurrence of SUA, warranting further investigation into the role of 4q structural variants in fetal vascular development.
In the present study, an additional 11 cases of pCNVs were detected using CMA, which were not identified by conventional karyotyping. This corresponds to an incremental diagnostic yield of 8.94% for CMA over karyotype analysis. The clinical significance of pCNVs in fetuses with SUA, however, remains a subject of ongoing debate. A meta-analysis conducted by Kim et al. [24], which included 124 fetuses with isolated SUA, reported no pathogenic CNVs, suggesting that isolated SUA may have one of the lowest rates of clinically significant CNVs among all ultrasonographic soft markers. Conversely, a more recent large-scale study involving 15,263 fetuses with various soft markers found that certain isolated markers, including SUA, thickened nuchal fold, mild ventriculomegaly, and absent or hypoplastic nasal bone, were associated with higher detection rates of pathogenic or likely pathogenic CNVs [26]. In the present cohort, an 8.0% pCNV detection rate was observed in the isolated SUA group, which aligns with findings from prior research [25].
Notably, the pCNVs identified in the present study did not exhibit a statistically significant enrichment pattern across specific genomic loci. Among the detected syndromes, Wolf-Hirschhorn syndrome (WHS) is particularly relevant due to its association with relatively nonspecific prenatal ultrasound features. However, SUA has been reported in approximately 44.4% of fetuses with WHS, suggesting it may serve as an early indicator of this condition [26]. Additionally, a previous study conducted by Kohlschmidt et al. [27] described cases involving partial monosomy 4 and partial trisomy 18 in a living male infant and a prenatally diagnosed male fetus, both of whom presented with a single umbilical artery, reinforcing the association between chromosomal imbalances involving chromosome 4 and SUA. Similarly, another report documented a fetus with partial monosomy 4p, who exhibited multiple prenatal anomalies including intrauterine growth restriction (IUGR), SUA, cleft lip and palate, and other soft marker anomalies [28]. Notably, in Case 3 of the present study, a microdeletion in the 4p16.3p16.1 region accompanied by a microduplication in 18q23 was identified in a fetus presenting with SUA, increased nuchal fold, and intrauterine growth restriction. These findings mirror a previously documented case of partial monosomy 4p, which was followed by pregnancy termination at the family’s request [28]. This report further implicates microdeletions at 4p16.3, the critical region for Wolf–Hirschhorn syndrome, in the development of SUA. A recent investigation [29] has linked 16p11.2 deletions with SUA. In the present cohort, Case 8 presented with both SUA and a 1.7 Mb deletion at 16p11.2. However, developmental assessment at one year showed normal growth and no obvious deficits, implying that SUA in this instance may not reflect full phenotypic expression of the 16p11.2 microdeletion. In Case 10, a microdeletion at Xq22.1 was detected in an isolated SUA fetus, marking the first such report. Although this points to a novel genomic region of interest, additional studies are required to determine whether this represents a causal relationship or a coincidental finding.
The detection of VOUS via CMA introduces substantial challenges in genetic counseling. Such ambiguous results can exacerbate parental anxiety and, in some cases, prompt termination of pregnancies that might otherwise yield healthy outcomes. Reliable interpretation of VOUS necessitates a multifaceted approach, including parental CMA testing, longitudinal detailed prenatal ultrasonography, and access to extensive population- and case-based databases to inform clinical decisions. In this cohort, five fetuses (4.07%) presented VOUS. Case 14 involved a 249.5 kb microdeletion at 7q36.1 inherited from the mother, who exhibited skeletal anomalies; however, no clinical issues were observed in the child over a five-year follow-up, suggesting that this deletion was unlikely to underlie the maternal phenotype. This example highlights the importance of prolonged surveillance and prudent interpretation of VOUS findings, while contributing valuable data to genetic counseling resources for similar future cases.
In the present study, 96 fetuses had normal CMA findings, and 94 of these pregnancies reached term. Among them, there were two stillbirths, three preterm deliveries, and one neonatal death shortly after birth. Earlier studies have proposed that SUA might impair placental circulation and nutrient transport, which could lead to outcomes like small-for-gestational-age infants and stillbirth [30]. The link between SUA and preterm delivery, however, remains debated. A study conducted by Mailath-Pokorny et al. [31] reported that isolated SUA is associated with an increased risk of preterm birth. In contrast, other findings suggest that isolated SUA does not elevate the risk of spontaneous preterm delivery [32]. In the present study, five pregnancies ended preterm, but without a matched control cohort, it was not possible to establish a causal relationship. Larger, controlled studies will be necessary to clarify whether SUA truly influences preterm birth rates.
In conclusion, in the present study, 123 cases of fetuses with SUA were identified among 5,014 pregnant women undergoing prenatal diagnosis in Southeast China. Several pCNVs were found to be potentially associated with SUA, including microdeletions in regions 4q34.3–q35.2, 4p16.3 (associated with Wolf-Hirschhorn syndrome), and 16p11.2. Additionally, the findings further support a correlation between long arm abnormalities of chromosome 18 and the presence of SUA. These results highlight the significant role of CMA in the etiological evaluation of fetuses with SUA, including those with isolated findings. CMA provides a higher resolution than conventional karyotyping and contributes valuable insights for prenatal genetic diagnosis. The present study offers additional evidence to inform clinical decision-making and genetic counseling for families affected by SUA.