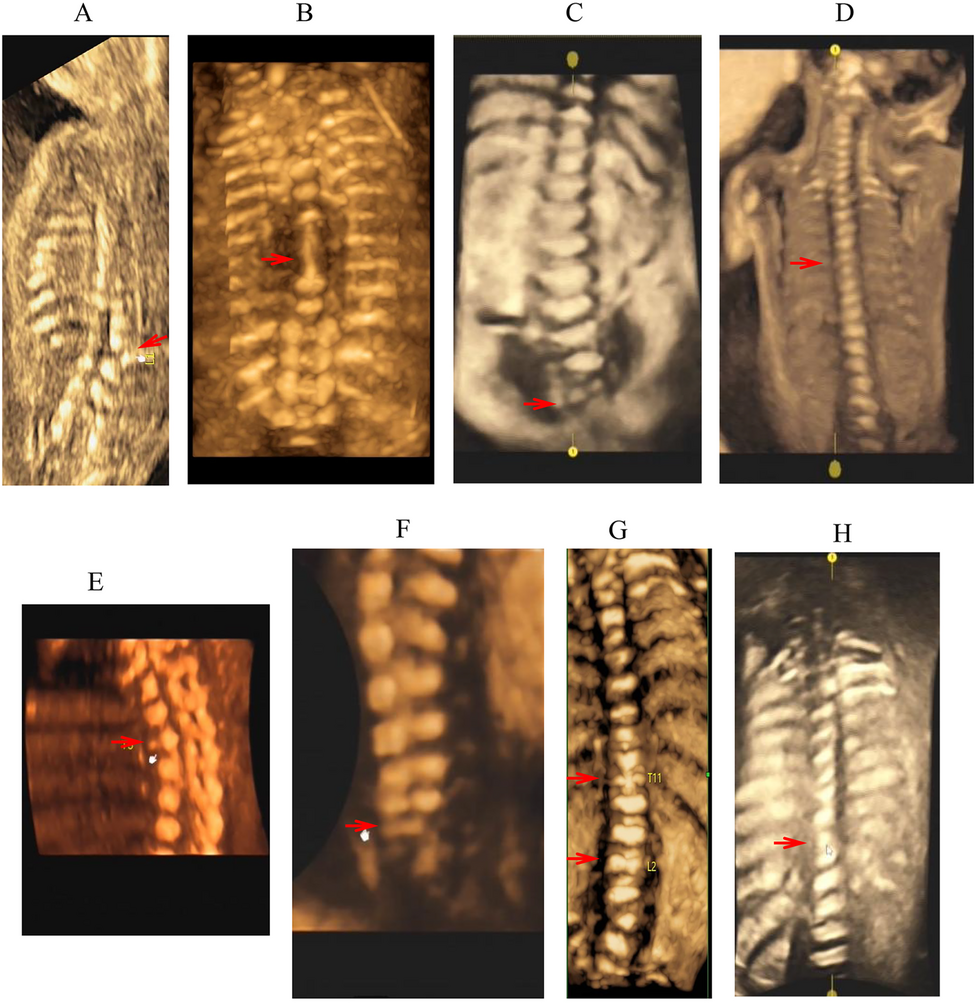
Examination of the fetal spine is an important part of routine prenatal ultrasound screening. The examination is significantly meaningful for prognosis prediction and birth defects control. However, due to our limited understanding of the etiologies and outcome of fetal vertebral defects, prenatal diagnosis based on the ultrasound findings is difficult [17, 18]. By reviewing the medical records at a tertiary hospital, a previous study from China reported on 71 cases of vertebral malformations detected by ultrasound among 69,013 fetuses, showing a positive rate of approximately one per 1,000 fetuses[19]. The majorities of these cases were hemivertebra (56.3%) and butterfly vertebrae (32.4%).
In an earlier retrospective study by Lemire et al., 66 fetuses with vertebral defects were identified prenatally by ultrasound over a 10-year period at a single center, with 48 (72.7%) fetuses having associated anomalies involving other systems[8]. A more recent study by Hu et al. reported on prenatal diagnosis and pregnancy outcome in 52 fetuses with vertebral abnormalities, of which 19(36.5%) were in complex with other anomalies[20]. In our study, 4 out of 12 cases (33%) were non-isolated, with three cases being congenital heart defects. Fisher’s exact tests showed that there was no significant difference in the ratio of non-isolated cases versus isolated cases between the two independent studies from China, while the two studies had a lower rate of non-isolated case compared to that (72.7%) of the earlier study (Table 2). The lower rates of non-isolated case in this case series and Hu et al.’s study might be explained by smaller sample sizes and late gestational ages, compared to those of the Lemire’s study (Table 2). Chromosome aberrations are a common cause of fetal vertebral anomalies. The ultrasound scan in early pregnancy has been shown to yield a high discovery rate for chromosome aberrations [21]. Furthermore, in recent years, the wide application of non-invasive prenatal testing (NIPT) or NIPT plus combined with ultrasound examination in early pregnancy has significantly improved the detection rate of common trisomies, rare chromosome aneuploidies and microdeletion/microduplication syndromes [22, 23]. This may contribute to a lower detection rate of non-isolated cases of fetal vertebral defects at later gestational ages.
Table 2 A review and comparison of studies on prenatal cases of vertebral defects
Currently, there is no clear consensus about genetic investigation for fetal vertebral defects detected by ultrasound [8]. To explore the etiology, either karyotyping, chromosomal microarray analysis, or both were previously applied to fetuses with prenatally identified vertebral defects in the earlier study conducted by Lemire et al., which yielded positive results in 3 (5.6%) out of the 53 cases tested, including two cases of karyotypic abnormalities (trisomy 13, trisomy 22) and one case of microdeletion (9q33.1-q34.11 deletion)[8]. More recently, He et al. conducted CMA on 52 prenatal cases with vertebral abnormalities and detected 7 cases of pathogenic CNV (n = 6) or aneuploidy (n = 1). Further, WES on 15 cases, among which 11 cases were with negative CMA results, led to diagnosis in additional three cases, involving the genes FLNB, KMT2D, and DLL3[20]. In this study, we identified three 16p11.2 deletions, one 7q36.1-q36.3 deletion using CMA as a first-line tool and a novel frameshift duplication mutation in TBX6 by WES for cases with negative results for pathogenic CNVs. As shown in Table 2, the more recent study by Hu et al. and our study overall had higher diagnosis yields than the earlier study by Lemire et al. This may be attributed to an increasing density of probes in oliognucleotide microarrays [24] and the new application of WES. Both our study and Hu et al.’s study highlight a crucial role of CMA in prenatal diagnosis of fetuses presenting vertebral malformations, accounting for 70%~80% of all the diagnosed cases. Furthermore, WES, giving a diagnosis in an additional 6–8% of cases, had a significant contribution to the total diagnostic yield in both studies. Interestingly, our data shows that the diagnostic yields are not statistically different between the isolated and non-isolated group, in line with the results of Hu et al.’s study[20]. Based on all these observations, it is suggested that CMA should still be used as first-line testing for fetuses presenting vertebral defects on ultrasound imaging, and WES can be offered when a diagnosis by CMA is not given.
It has been demonstrated that heterozygous 16p11.2 microdeletion (including the TBX6 gene) in trans with a hypomorphic allele defined by three SNPs (the T-C-A haplotype) is a common cause for congenital scoliosis in postnatal cases[12, 14] and for vertebral malformations in prenatal cases[25]. Our findings further support the prevalence of the recurrent 16p11.2 deletion in fetuses with vertebral anomalies. Of note, apart from CNV, our study also shows that heterozygosity of a TBX6 frameshift variant and the T-C-A haplotype on the opposite allele can cause fetal vertebral anomalies. Our investigation gave a total diagnostic rate of 41.7% (5/12), which is not statistically different from the total diagnostic yield (19.2%, 10/52) of the study by Hu et al. (Table 2). Both of the studies suggest a feasible strategy for genetic testing on prenatally identified vertebral anomalies.
TBX6 as a transcription factor plays an important role in somitogenesis[11]. Patients with TBX6-associated congenital scoliosis (TACS) are genetically characterized by a null/loss-of-function mutation in trans with the T-C-A haplotype on the other allele [12, 26, 27]. Haploinsufficiency of TBX6 is insufficient to cause congenital scoliosis, and thus a gene dosage model has been proposed [12]. Previous studies have examined a series of frameshift/nonsense/missense variants of TBX6 and demonstrated various severity of function loss due to nonsense mediated mRNA decay, decreased transcription activity, or cellular mislocalization[14, 27]. The present study identified a novel frameshift variant of TBX6, namely the c.989_990dup (p.(G331Pfs*168)) variant, which is thought to produce a disrupted TBX6 protein with the intact T-box domain. It can be inferred that the pathogenicity of the c.989-990dup variant may be attributed to a significantly decreased transcription activity of the encoded protein, similar to the c.935_936insGA (p.D312Efs*187) variant as shown in a previous study [14]. Heterozygosity for the c.989_990dup variant and a hypomorphic T-C-A allele may result in insufficient gene dosage of TBX6, therefore being responsible for the vertebral malformation phenotype in our study.
Our study is not without its limitations. Though our findings provide valuable insights into the genetic features in fetuses presenting vertebral defects, the sample size of the case series was small, and three of the 12 cases declined or were lost to follow-up, leading to incomplete information about pregnancy outcome and interpretation of decision making.