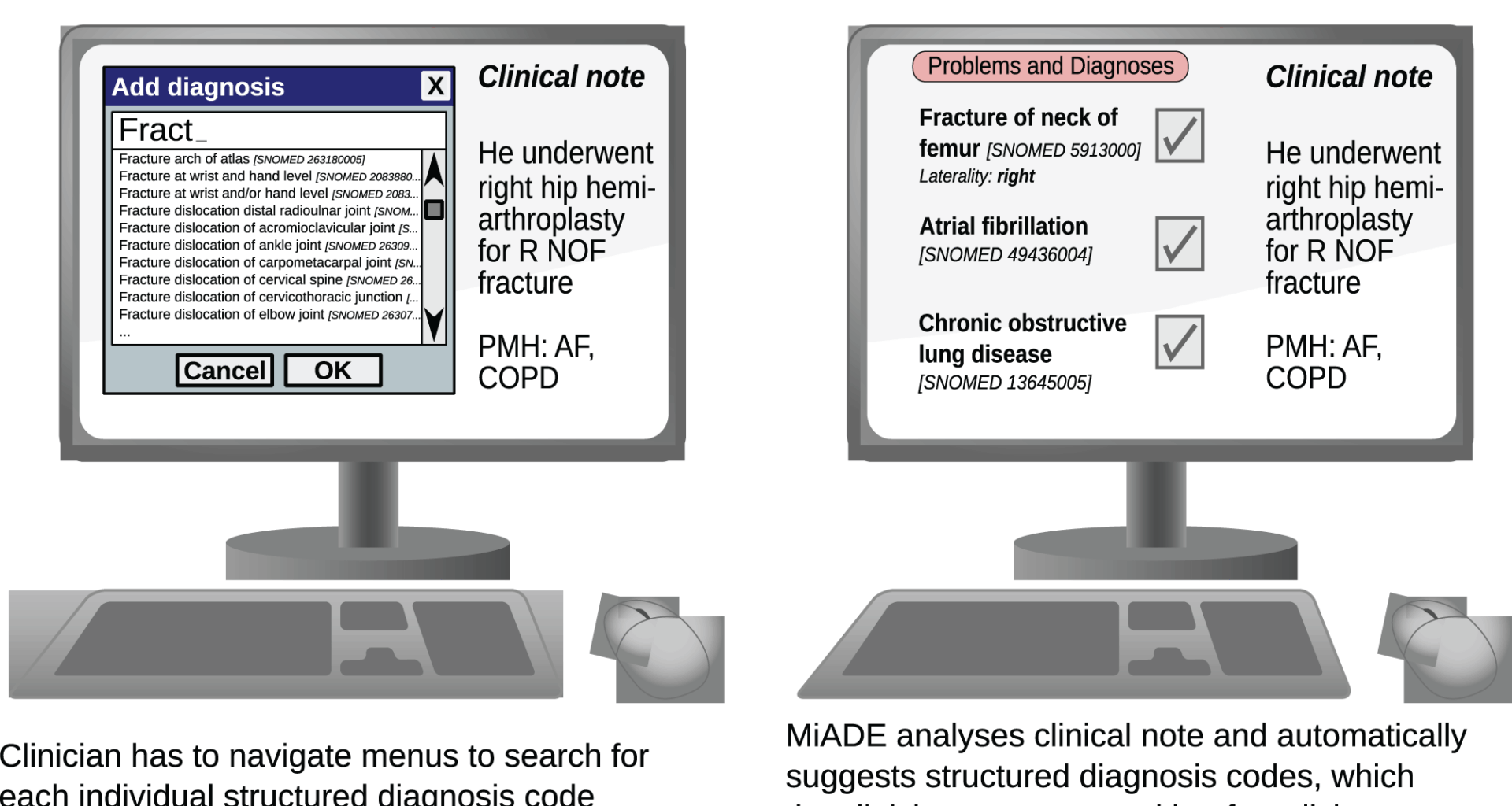
We developed a modular, open source NLP system called MiADE which is able to communicate with an EHR and process clinical notes in real time, returning suggestions to the EHR for display to the clinician. The clinician can use the EHR user interface to accept or reject the MiADE suggestions and reconcile them with existing information.
MiADE uses the open source MedCAT program [17], part of the Cogstack family of NLP tools [18], as its named entity recognition (NER) tool. MiADE does not have its own user interface but communicates with an EHR using data standards Health Level 7 Clinical Document Architecture (HL7 CDA) messaging, and can be readily adapted to use Fast Healthcare Interoperable Resources (FHIR). At UCLH, the user interface consisted of the ‘NoteReader’ component of the Epic EHR.
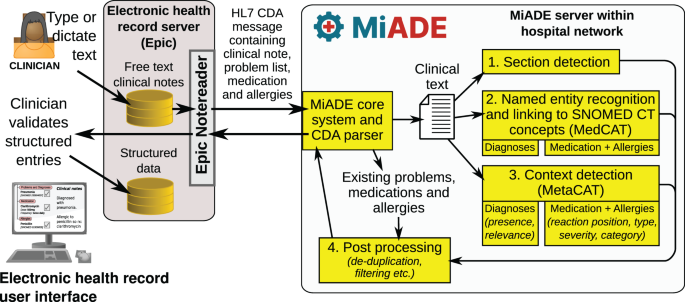
The system architecture is shown in Fig. 2, which illustrates how the key functions of the system link together:
(1)
Section detection
(2)
Named entity recognition and linking to SNOMED CT concepts (MedCAT)
(3)
Context detection (MetaCAT)
(4)
Post-processing
Schematic showing the overall architecture of the MiADE system
Section detection
While the prose of clinical documents is hugely variable, we found that clinicians often write diagnoses, problems or medications in list-like formats within sections with a meaningful header. The context of information within such sections is much easier to process than in prose. We therefore designed a text cleaning and paragraph detection process which can detect section headings based on a configurable set of regular expressions (regex). Annotated outputs are then post-processed according to the paragraph type, and only relevant concepts are returned.
Named entity recognition and linking to SNOMED CT concepts (MedCAT)
MiADE uses the open source MedCAT [17] library as its core NER algorithm. MedCAT is a self-supervised machine learning algorithm for extracting concepts using any concept vocabulary, available as an open source python package. MedCAT demonstrated superior performance on extraction of Unified Medical Language System (UMLS) concepts compared to SemEHR [19], Bio-YODIE [20], cTAKES [21] and MetaMap [22]. MedCAT models are trained unsupervised on a large text corpus to learn the context surrounding concept mentions in order to perform disambiguation. Supplementary supervised training can be carried out to help MedCAT learn additional synonyms and positive and negative examples of context. We have an active collaboration with MedCAT developers, and UCLH has already deployed MedCAT for retrospective clinical research [18].
MedCAT concept database and training
We built and trained separate models with different concept databases (CDB) for problems and medications / allergies, to allow each algorithm to be developed and implemented independently. We used a set of R scripts and the “Rdiagnosislist” R package to process SNOMED CT dictionaries and prepare the concept databases [23], using the UK SNOMED CT May 2022 version. The scripts and data are available in the miade-datasets GitHub repository (https://github.com/uclh-criu/miade-datasets/).
The problems concept database included symptoms, diagnoses, and important aspects of social history (e.g. care needs and housing), but not family history concepts, allergy concepts, normal findings or high-level concepts that are not relevant for a problem list, such as ‘Disease related state’. Acronyms expressed in SNOMED CT concept descriptions and a small number of common synonyms were added from a manually curated list.
The medication and allergy concept database included substances, symptoms that could be manifestations of allergic reactions, and medications from the Dictionary of Medicines and Devices (dm+d) [24] that are Virtual Therapeutic Moieties (VTMs) or Virtual Medicinal Products (VMPs).
MedCAT models are trained in unsupervised and supervised steps [17]. The unsupervised training step takes in a corpus of free-text documents and the CDB and learns the vector embedding representation of the context of each concept. This context representation is used to differentiate concepts when an ambiguous term is detected. For this step, we used a corpus of 800,000 UCLH clinical notes (mostly progress notes and discharge summaries) from April 2019 onwards (which is when the Epic EHR was installed at UCLH).
The supervised training process involves manual annotation to “teach” MedCAT to avoid incorrect annotations (i.e. words that do not have the same meaning as the concept they are linked to), and to learn additional correct synonyms and context. MedCAT can be trained to improve its performance in a particular domain of interest.
We chose discharge summaries as the initial focus for this project because of the clinical priority to improve handover between hospital and primary care, and to improve clinical coding and reimbursement. We therefore extracted a random subset of 400 discharge summaries for training from the UCLH corpus described earlier. These documents were annotated with SNOMED CT concepts by three clinically trained health informaticians (R.W., J.W., M.M.N.S.D.), using the MedCATtrainer tool provided by Cogstack [25], following a set of annotation guidelines (See Supplementary Text). We extracted a separate random subset of 50 discharge summaries for testing, which underwent double independent annotation. In cases of disagreement, annotations were reconciled manually by a third annotator.
Context detection (MetaCAT)
MedCAT includes the ability to train small bidirectional long short-term memory (Bi-LSTM) neural network models to detect the context of concepts that are recognised. These are known as MetaCAT models, and can be trained using the same annotated data as the supervised training step of the MedCAT model. MetaCAT models are trained with standard neural network training procedures and evaluated with precision, recall, and F1 scores.
We trained MetaCAT models for the Presence, Relevance and Laterality of problems. For medications and allergies, we trained models to classify the position of a reaction mention relative to the substance ReactionPos, whether a substance is an allergen or a medication that is being taken SubstanceCategory, whether an adverse reaction risk is an allergy or intolerance AllergyType, and Severity of allergies (see Table 1). We carried out the training with the concept of interest (problem, medication, substance or reaction) replaced by a generic placeholder token, to prevent the models from incorrectly inferring context from the concept itself (e.g. to avoid assuming a concept is suspected because it frequently appears suspected in the training data).
Table 1 Meta-annotations for different models
Outputs from the MetaCAT model are extracted into a MetaAnnotation class and returned as an attribute to the main concept. MiADE then applies a set of logic rules to the meta-annotations in combination with the section type to decide the final context of a concept. For example, if a concept is detected as “irrelevant” in a “previous medical history” paragraph, the meta-annotation is converted to “historic”, as the section heading takes precedence over the meta-annotation. Text without section headings is detected as “prose” paragraphs, and MiADE can either return or ignore concepts detected in prose paragraphs depending on the configuration setting structured_list_limit. This parameter limits the number of concepts that may be returned from prose. In the UCLH implementation it was set to −1 to ignore all concepts in prose, based on clinician feedback that it was preferable to avoid returning too many irrelevant concepts.
Synthetically augmented data for training MetaCAT models
One challenge we encountered in training the MetaCAT models was imbalanced classes in the annotated dataset: annotations of “suspected”, “historic”, and “irrelevant” occurred much less frequently than those of “confirmed” and “present”. To address this, we created additional synthetic training data using specially curated patterns of common phrases for expressing negation, suspected conditions, historic conditions, and allergies to augment the annotated dataset.
To facilitate the transparency and reproducibility of the training process with the synthetically augmented dataset, a streamlit dashboard was created to manage the dataset and model training process. This provides the ability to interactively review and modify the quantity of annotated data and synthetic data required in the training dataset. It can also automatically balance the class weights, version training parameters, and quickly visualise confusion matrices against the test set. The dashboard has been incorporated in the MiADE package.
Post-processing
The core MiADE pipeline contains the NoteProcessor, Annotator, and DosageExtractor modules. The pipeline takes in a Note object and returns a list of Concepts. Functionalities to clean text and detect paragraph section headings are provided within the Note object. NoteProcessor acts as the main Application Programming Interface (API), to which the implementer can add MedCAT models to using the .add_annotator() method. The Annotator class wraps a single MedCAT model and processes the outputs from the model using custom post-processing algorithms we have developed. For usage with medication concepts, a DosageExtractor module is also provided, which extracts medication dosages and converts the output to CDA-compatible format. MiADE performs de-duplication of all concepts returned.
The MiADE processing pipeline is open source and available for use with different MedCAT models. To initialise MiADE, the implementer needs to provide a directory of MedCAT models, trained to extract concepts of different types (problems, medications, and allergies) and fine-tuned on local text datasets. Post-processing components are configurable via a set of lookup dictionaries for the conversion steps described below, as well as the regex that are used to extract section headings. A filter list can also be specified to blacklist irrelevant concepts, which can be amended without having to retrain the MedCAT model. The concept database and lookups used in our MiADE study are available on the public ‘miade-datasets’ GitHub repository [26].
Depending on need, MiADE pipeline components can be disabled in the configuration file e.g. for a use case that does not require certain post-processing steps such as dosage extraction or medication conversions. Implementers are therefore able to create plug-and-play processing pipelines with MedCAT models.
Post-processing of problems
Concepts detected with “negated”, “suspected”, and “historic” contexts are converted to corresponding SNOMED CT terms via lookup dictionaries, e.g. fever with a “negated” context is converted to the concept apyrexia. Concepts without conversion matches and irrelevant concepts are filtered out. Our implementation converts historic procedures to “history of” concepts (e.g. ‘History of coronary artery bypass graft’) but leaves historic diagnoses unchanged. This behaviour can be easily configured by custom lookup tables.
Post-processing of medications and allergies
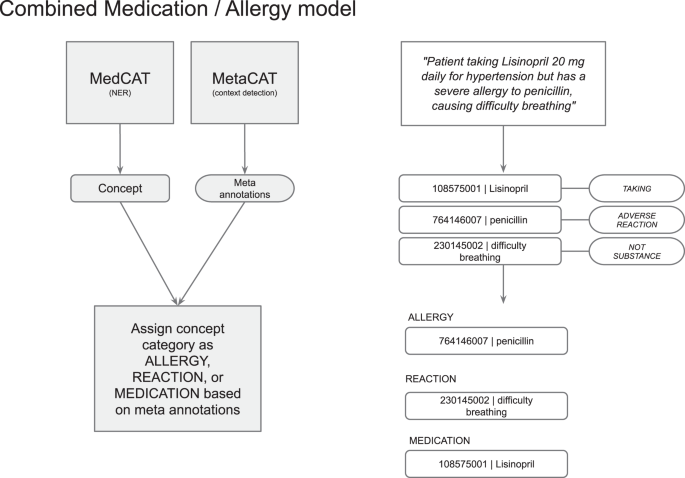
The default MiADE algorithm extracts medications and allergies from a single MedCAT model, then differentiates and assigns medication and allergy type labels based on meta-annotation results.
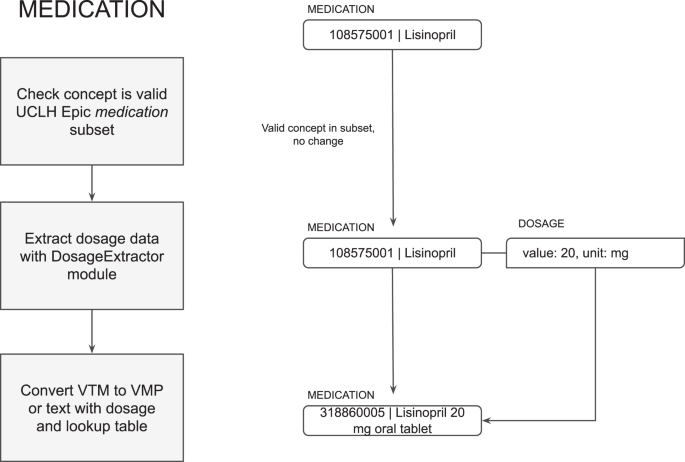
First, the detected concept is processed as either a medication (“taking”), allergy (“adverse reaction”) or not a substance (“not substance”, such as a reaction) based on the meta-annotations. The concept is then validated and converted to a SNOMED CT subset in each category if appropriate (see Fig. 3). This can be configured based on different implementation and use cases.
NER of medication and allergy model with example workflow
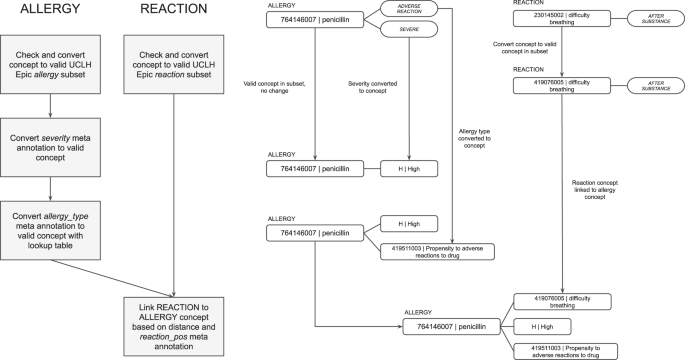
MiADE then performs linkage of the allergen to the allergy type, severity, and reaction. A single allergy entry is returned as an allergen SNOMED CT concept linked to concepts for allergy type, severity, and reaction in the CDA sent to the EHR (see Fig. 4). For each allergen substance detected, the severity is extracted through MetaCAT models and converted to appropriate SNOMED CT codes. The type of adverse reaction risk is also extracted through the MetaCAT model as either “intolerance”, “allergy”, or “unspecified”. An overall SNOMED CT concept for the allergy record type (e.g. “Food intolerance”) is chosen based on the adverse reaction risk type and substance type. Lastly, the reaction is detected by the core MedCAT model and linked to the closest allergen concept, based on whether the meta-annotation for the reaction concept is “before substance” or “after substance”.
Post-processing of allergy concepts with example workflow
Medications are detected either as Virtual Medicinal Products (VMP) if the dose form is stated, or Virtual Therapeutic Moieties (VTM) if only the substance is stated. The dosage is detected using the DosageExtractor algorithm described in the next section. VTMs may not be valid for return to the EHR depending how the EHR drug dictionary is set up, so MiADE includes a conversion table to convert them to VMPs for common doses of oral tablet medications. If the conversion is not possible, they are returned as text, in which case the clinician will need to select an appropriate VMP manually when reconciling the NLP output. Dosages are returned as CDA-compatible dosage information, where available (see Fig. 5).
Post-processing of medication concepts with example workflow
Extraction of medication dosages
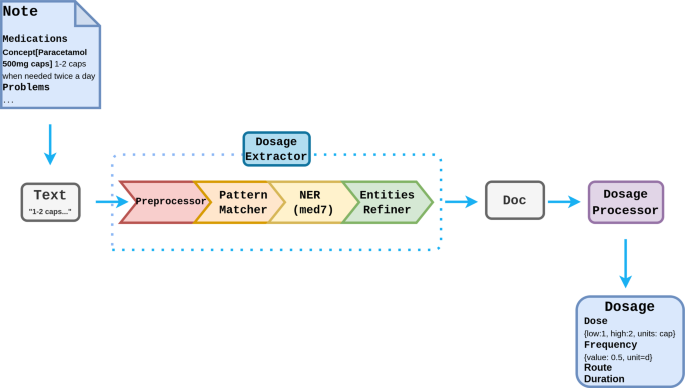
The first stage in dosage extraction is to identify words and phrases that constitute components of the dose. This is carried out using the med7 NER model, a spaCy neural network-based named entity recognition system for prescription components, trained on MIMIC III [27]. A rule-based algorithm was developed based on the CALIBERdrugdose R package [28] to calculate structured medication dosages. The dosage extractor is built as a spaCy pipeline and can be configured through the main MiADE API or used independently (See Fig. 6).
Pipeline design for drug dosage extraction
Pre-processing
The pre-processing steps are adapted from CALIBERdrugdose algorithm [28], which involves using two lookup dictionaries (singlewords and multiwords) to standardise words and expressions in the text and correct common spelling mistakes. This is followed by a NumbersReplace step to standardise the way quantities are expressed in the text (e.g. “2 ×200 mg” becomes “400 mg”, “1/2” becomes “0.5”).
Pattern matching
The Pattern Matcher step matches structural sentence patterns and performs rule-based entity tagging to disambiguate dosage information within the text, particularly dosage instructions that follow a specific format e.g. “1 tablet to be taken twice a day, 30 tablets”. It then uses the patterns lookup table from CALIBERdrugdose to extract dosage results and stores the results as an attribute in the spaCy Doc object.
Named entity recognition
The Med7 model is used to extract the labels: Dosage, Drug, Duration, Form, Frequency, Route, Strength. As not all labels extracted by Med7 are needed, some rule-based refinements are made to the NER results:
Consecutive labels with the same tags are combined
Drug and Strength labels are ignored (detected as part of VTM / VMP concepts by the main algorithm)
Strength labels are combined with Dose labels if they are consecutive
Drug labels are relabelled Form if they follow Dose (drug should be in concept name and should not appear in dose string)
Composition of a CDA dosage class
The processor takes the lookup and NER results from DosageExtractor and parses them into the CDA-compatible Dosage class:
Dose [quantity, unit, low, high]
Frequency [value, unit, low, high, standard_deviation, institution_specified, precondition_asrequired]
Duration [value, unit, low, high]
Route [code, displayName]
Testing
We tested the following aspects of MiADE before deploying it in UCLH. First, we tested the accuracy of the MedCAT NLP models for detecting SNOMED CT diagnosis concepts, to ensure that the model training was adequate. Second, we carried out user acceptance testing of the complete pipeline integrated with EHR to ensure that it was usable by clinicians and behaving as expected. Third, to verify that the system can potentially save time for clinicians, we compared the time taken to enter a problem list with and without MiADE for simulated patients.
It was not possible to deploy medications and allergies initially because of issues with the EHR integration, hence model testing has not yet been performed for this functionality. Additional work is required to validate the medication and allergy pipeline before it is ready for clinical use.
Performance of the problems MedCAT model
We assessed the performance of concept detection for problems (symptoms and diagnoses) by testing the MiADE MedCAT model on a gold standard test set of 50 randomly selected discharge summary notes from UCLH, across a range of clinical specialties. These notes were double annotated by health informaticians and then manually reviewed to resolve any discrepancies.
We evaluated the MiADE problems algorithm on three components: concept detection, context detection, and full pipeline performance. The concept detection refers to the NER performance of MiADE, assessing the number of concepts detected irrespective of the metadata returned from the context detection algorithms. Context detection for problems ascertains affirmation status (affirmed, negated or suspected), temporality and relevance. The full pipeline performance assesses whether MiADE correctly suggests a concept given the full pipeline of concept detection and applying post-processing and filtering logic based on the metadata from context detection. For each component we quantified the performance with precision, recall, and F-1 score. For all evaluations we disabled the paragraph detection and deduplication components to assess the performance of the MedCAT / MiADE algorithm on interpreting prose. Each component was evaluated on two filtered subsets of concepts – ‘Symptoms + Diagnoses’ (concepts with the SNOMED CT semantic types “Finding” or “Disorder”, omitting administrative concepts), and ‘Diagnoses Only’ (mostly SNOMED CT “Disorder” concepts, with a few clinically important findings such as “Seizure” included).
The concept detection and full pipeline performance were evaluated on a per-document basis, without positional matching of concepts. A SNOMED CT concept that was detected by MiADE but not present in the gold standard is classed as a false positive, and vice versa for false negative.
To evaluate context detection, we needed to match up concepts in the MiADE output with those in the annotated gold standard. We were unable to do this directly because of shifts in the concept spans from the text cleaning and pre-processing stages. Instead, for each document we first cross-joined the set of concepts returned by MiADE with the gold standard set of concepts. We then used a custom fuzzy match algorithm based on a combination of positional matching and string similarity to identify spans that corresponded to the same concept. From this set we checked if the SNOMED IDs of each matched concept matched, giving the final set of correctly detected and suggested concepts. The context detection evaluation did not include concepts that were identified as false positives, false negatives, or were not matched by the fuzzy-matching algorithm.
User acceptance testing
MiADE was installed on a server linked to a test installation of Epic. Clinician investigators working on MiADE tried out the system with simulated patient histories to ensure that the user interface worked as expected. This was essential because the manner in which Epic renders CDA data is frequently lacking in comprehensive documentation, necessitating a process of trial and error to verify that the information is displayed as intended.
Testing speed of data entry
We set up a data entry task for clinicians and timed them using the default EHR method of entering SNOMED CT concepts (using the EHRs terminology browser) and using MiADE (where the problems would be auto-suggested for clinician verification; see Fig. 1). Clinicians were asked to perform the task on a set of 20 simulated clinical notes each containing a problem list with 10 problems. The problem names were SNOMED CT synonyms for diagnosis concepts randomly selected from the frequency distribution of SNOMED CT concepts in UCLH problem lists in 2019–2021. This task was designed to ascertain the maximal possible time saving using MiADE, as the terms were SNOMED CT names without variations or mis-spellings and would be accurately detected by MiADE.
Study participants were five clinicians who had experience of using Epic, with a range of seniority and familiarity with digital technology.
Integration with EHR
We deployed and integrated MiADE into the Epic EHR system at UCLH. In addition to the core open source MiADE library, we also developed a server and a CDA parser for integration into EHRs. The MiADE server is deployed as a Docker container on hospital internal network and communicates with Epic over secure HTTPS. It implements the Simple Object Access Protocol (SOAP) method ProcessDocument, which is invoked by Epic to send over a CDA document that contains the note entered by the user in Epic NoteReader. To trigger this request, the user selects or toggles the “Send to NoteReader” button when they are in the Epic NoteReader interface. The input CDA is parsed and processed by the MiADE server, and a response CDA document containing the extracted data is returned to Epic.
We implemented the SOAP endpoint invoked by Epic as a Web Server Gateway Interface (WSGI) sub-application mounted to a FastAPI app at /notereader/, so that the server is able to receive both RESTful and SOAP requests. This modularisation lends extensibility to the server implementation and allows possibility of integration with different EHR systems, including the option to receive and return data in FHIR format.
Pathway to deployment
MiADE was initially incorporated into a test environment of Epic at UCLH, which is not linked to the live clinical system and does not include actual patient data. Prior to production deployment of MiADE, we load tested our servers using the load testing framework locust for a maximum capacity of 100 concurrent users to ensure the server is able to support the load of the feasibility trial. The server was distributed across 8 worker nodes (one for each CPU core), with load balancing handled by nginx. Test request data used were CDA document requests with varying lengths of structured data and progress note lengths.
We developed a clinical safety case following the NHS digital safety standards [29, 30]. We performed a risk evaluation by convening hazard workshops with clinicians, health informaticians and software engineers. A hazard log was compiled based on the workshop discussions. Risks fell into three broad themes: risks due to failure of the MiADE software (crashes/data leaks etc.), risks due to the effect of the integration of MiADE on the EHRs (uncovering bugs in an otherwise unused function of the EHRs), and clinician behaviour change (failure to properly vet suggestions from MiADE).
We worked with the EHR specialist team at the hospital to carry out a series of tests to validate that the communication between the EHRs and MiADE was working, data was correctly placed into the medical record, and that the interface functioned as expected.
Following testing and hazard workshops, a safety case was submitted to the Trust Clinical Safety Committee, and subsequent Trust approvals were sought for live deployment within the context of an evaluation study for up to 100 users. The initial live implementation was limited to diagnosis only due to unresolved issues with the Epic EHR integration for medications and allergies.