 Credit: Olena_T / Getty Images
Credit: Olena_T / Getty Images
Antimicrobial resistance (AMR) is one of the world’s most urgent health challenges, with the WHO’s recent report showing there are “too few antibacterials in the pipeline” and the limited commercial incentives deter investment in antibiotic discovery.
The methylenomycins (methylenomycin A and B) are antibiotic compounds—produced by the bacterium Streptomyces coelicolor A3—that are cyclopentanone derivatives. They are effective against both Gram-negative and Gram-positive bacteria.
In a new study published in the Journal of the American Chemical Society entitled, “Discovery of Late Intermediates in Methylenomycin Biosynthesis Active against Drug-Resistant Gram-Positive Bacterial Pathogens,” researchers report on a new antibiotic pre-methylenomycin C lactone—an intermediate chemical in the natural process that produces methylenomycin A.
“Methylenomycin A was originally discovered 50 years ago and while it has been synthesized several times, no one appears to have tested the synthetic intermediates for antimicrobial activity,” notes Greg Challis, PhD, professor in the Department of Chemistry at the University of Warwick, and Biomedicine Discovery Institute at Monash University. “By deleting biosynthetic genes, we discovered two previously unknown biosynthetic intermediates, both of which are much more potent antibiotics than methylenomycin A itself.”
When tested for antimicrobial activity, one of the intermediates, pre-methylenomycin C lactone, was shown to be over 100 times more active against diverse Gram-positive bacteria than the original antibiotic methylenomycin A.
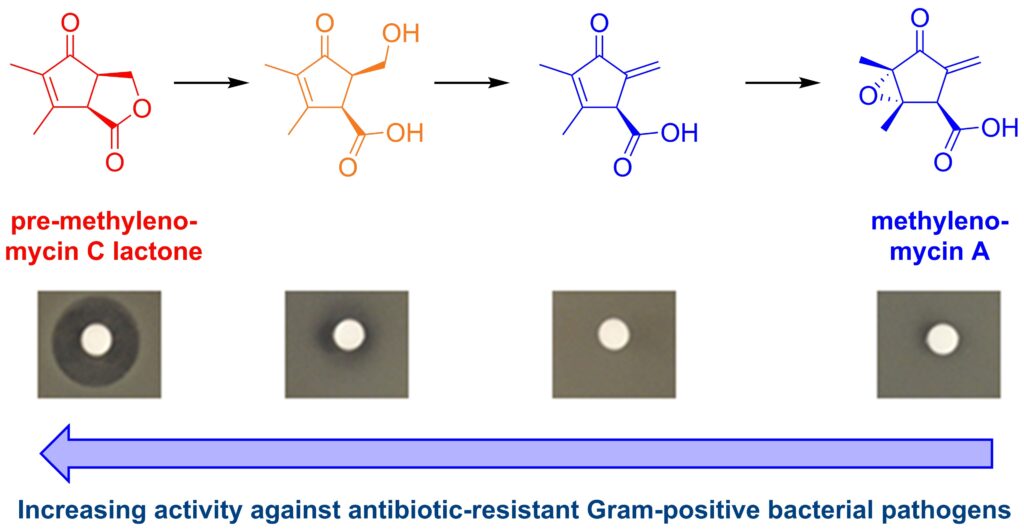 The biosynthesis pathway of methylenomcyin A as discovered in this experiment. The new precursors, especially pre-methylenomycin C lactone, show increased potency against Gram-positive bacterial pathogen compared to the original methylenomycin A. [Greg Challis/University of Warwick]Specifically, it was shown to be effective against Staphylococcus aureus and Enterococcus faecium, the bacterial species behind MRSA and VRE respectively.
The biosynthesis pathway of methylenomcyin A as discovered in this experiment. The new precursors, especially pre-methylenomycin C lactone, show increased potency against Gram-positive bacterial pathogen compared to the original methylenomycin A. [Greg Challis/University of Warwick]Specifically, it was shown to be effective against Staphylococcus aureus and Enterococcus faecium, the bacterial species behind MRSA and VRE respectively.
“Remarkably, the bacterium that makes methylenomycin A and pre-methylenomycin C lactone—Streptomyces coelicolor—is a model antibiotic-producing species that’s been studied extensively since the 1950s. Finding a new antibiotic in such a familiar organism was a real surprise,” notes Lona Alkhalaf, PhD, assistant professor, University of Warwick. “It looks like S. coelicolor originally evolved to produce a powerful antibiotic (pre-methylenomycin C lactone), but over time has changed it into methylenomycin A—a much weaker antibiotic that may play a different role in the bacterium’s biology.”
Importantly, the researchers could not detect any emergence of resistance to pre-methylenomycin C lactone in Enterococcus bacteria under conditions where vancomycin resistance is observed. Vancomycin is a “last line” treatment for Enterococcus infection, so this finding is especially promising for VRE, a WHO High Priority Pathogen.
The next step in the development of the antibiotic will be pre-clinical testing. In a coordinated publication earlier this year in the Journal of Organic Chemistry, the work reported on a scalable synthesis of pre-methylenomycin C lactone, paving the way for further research.
“This discovery suggests a new paradigm for antibiotic discovery,” Challis asserts. “By identifying and testing intermediates in the pathways to diverse natural compounds, we may find potent new antibiotics with more resilience to resistance that will aid us in the fight against AMR.”