Investigating genetic relationships and diversity within plant varieties is essential to promote the sustainable use of genetic resources29. Although other studies have provided global or Africa-centric insights into cowpea diversity4,9,38,39,40,43,64,65, this study fills a major gap by focusing on Asian genetic resources collected from 18 countries across multiple continents. Only a few studies have investigated the detailed genetic diversity of Asian cowpea genetic resources.
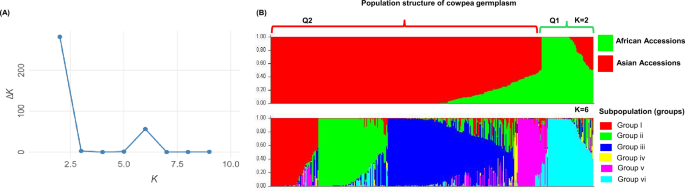
Analyzing population structure is crucial in identifying the genetic foundation of complex traits for association studies66. In this study, we examined 410 cowpea accessions selected for their agronomic trait diversity, specifically seed coat color and pod length, representing genetic variation across 18 countries. Our analysis of population structure revealed that NARO cowpea germplasm is genetically divided into two geographically distinct subpopulations. The first subpopulation (Q1) includes accessions primarily from West Africa, Northeast Africa, and South Asia, while the second subpopulation (Q2) comprises accessions from East Africa, South Asia, Southeast, East Asia, and Europe. At K = 2, the analysis identified two main gene pools as indicated by the peak of delta K (∆K), highlighting clear genetic differentiation within the cowpea population. Further analysis at K = 6 revealed additional sub-structuring, dividing the accessions into six distinct groups, likely representing sub-division events among the three cultivated subspecies: grain, fodder, and vegetable cowpeas.
The Q1 subpopulation contained two subspecies: Vigna unguiculata ssp. unguiculata, which is cultivated for seeds in sub-Saharan Africa and exhibits white, brown, and black seed coats with pod lengths of 10–22 cm, and Vigna unguiculata ssp. cylindrica, which features black and red seed coats. Q2 consisted of accessions from ssp. unguiculata and ssp. sesquipedalis, with the latter distinguished by its red, black, brown, and purple seeds and pod lengths (30–50 cm).
The levels of admixture varied between the two subpopulations, with Q1 showing 5.4% admixture and Q2 displaying 14.8%, suggesting potential gene flow or shared ancestral origins. The mixed geographical distribution of these genetic groups was further supported by findings from the DAPC and BIC analyses. The higher level of admixture in Q2 is likely influenced by historical and breeding-related factors. As cowpea spread from Africa to Asia, it diversified through natural and human-mediated selection, leading to the development of ssp. sesquipedalis, an early maturing cultivar valued for its long and tender pods used as vegetables. Asia’s long history of cowpea cultivation, combined with traditional farming practices, seed exchange networks, and farmer-driven selection, facilitated genetic mixing among landraces and cultivars. Additionally, modern breeding programs and germplasm exchange between regions have further contributed to admixture by incorporating diverse parental lines for cowpea improvement.
The geographical distribution of the identified populations was consistent with the results obtained from the PCA and phylogenetic tree based on genetic distance analysis. The delineation of accessions in Q1 primarily consisted of those from Western and Northeast Africa, along with East, South, and Southeast Asia. In contrast, Q2 included accessions from East, South, and Southeast Asia, East Africa, and Europe, and these results remained consistent across multiple analyses. Notably, West African accessions were predominantly grouped in Q1, with minimal representation in Q2, except for a single Nigerian accession. This distinct clustering suggests a unique genetic signature or ancestry for West African accessions compared to other regions.
While no population was entirely composed of accessions from the same region, certain regions showed a strong tendency to fall into either group Q1 or Q2. West African accessions (Benin and Ghana), Southeast Asia (Laos, Malaysia, and Myanmar), and Chinese accessions, predominantly clustered within a single group, indicating admixture ancestry. The clustering of accessions into Q1 or Q2 suggests a complex interplay of genetic diversity, history of genetic exchange, migration, and admixture among the studied populations. Most cowpea accessions exhibited mixed parental lines, particularly those from West Africa, as well as a single accession from Papua New Guinea, except for those from Sudan, Russia, and Thailand. The Papua New Guinea accession classified as ssp. sesquipedalis was identified as a breeding line and remained unassigned to a specific cluster due to its mixed genetic background. This suggests a potential historical or breeding-related exchange from Asia. The Russian accession belonged to Q2, while Sudanese and Thai accessions were found in Q1 and Q2. However, the limited number of accessions contributed from Russia and Sudan (one to five) may not adequately capture the full diversity of cowpea in these areas.
Compared to previous studies analyzing global cowpea germplasm collections, which identified genetic divergence between West and East African germplasm9,61,62,64, our findings reinforce this distinction as the primary structuring factor in cowpea diversity. Despite the limited number of East African representatives, Ethiopian accessions grouped in Q2 alongside Asian accessions, suggesting a genetic similarity between these regions. The confinement of West and Northeast African accessions to Q1, along with evidence of a mixture between West African accessions and Asian accessions, further underscores recently shared ancestry lines between these populations. Among the 347 Asian accessions, 79.3% and 6.6% were assigned to Q2 and Q1, respectively, indicating long-term coexistence of both genetic backgrounds in Asia. Importantly, the clear distinction between cowpea accessions from Asia and Europe and those from West Africa remains a significant and consistent finding.
Further analysis using DAPC and BIC identified six main groups (1 to 6), which were further divided into 13 subclusters, reflecting geographic influences. At K = 6, the scatter plot distinctly illustrates divergence among these identified genetic clusters. Group 1, the most diverse, contained 8 subclusters, indicating significant genetic variation likely due to its broad geographic distribution and extensive genetic exchange. Group 2, in contrast, comprised a single, more homogenous cluster predominantly associated with the Asian gene pool (Q2). Groups 3, 4, and 6, each containing one subcluster, showed limited genetic variation and were primarily associated with the African gene pool (Q1). Group 5 displayed admixed genetic profiles, suggesting gene flow between populations. Within the Asian gene pool, DAPC analysis confirmed a distinct subcluster of ssp. sesquipedalis and other unique subclusters for Nepalese and Japanese accessions. The distinct subcluster of Nepalese is likely attributed to the diverse and extreme environmental conditions in the Hindu Kush Himalaya Range, which promotes adaptation and genetic differentiation in crops like cowpeas66,67,68. The genetic divergence found in Nepal and Japan may result from distinct climates. Nepal’s high-altitude, subtropical, and temperate conditions, and Japan’s cooler, high-precipitation environment, may have shaped unique stress-adaptive traits in cowpea from these regions. Additionally, Nepal and Japan both preserve landraces, however, Japan also emphasizes targeted breeding, further reinforcing genetic distinctiveness. The high-altitude and high-precipitation environments are distinct from the area of domestication in Africa, suggesting that Nepalese and Japanese genetic groups may harbor environmentally adaptive alleles.
Despite the identified subclusters, the genetic variation within the African population was low to moderate, suggesting that geographical proximity and exchange of genetic materials within West African regions have reduced genetic isolation. Although no single group exclusively comprised accessions from the same country or region, South Asian accessions showed a tendency to cluster together, reinforcing regional genetic coherence, consistent with phylogenetic tree groupings. The low to moderate genetic variation observed, particularly within the African population, where genetically similar individuals are clustered into distinct gene pools, may limit the availability of novel alleles for cowpea breeding programs. Consequently, crossing members of different populations, such as African and Asian accessions, is expected to enhance genetic diversity and produce more valuable progenies. As Huynh et al.61 previously noted, breeding programs often operate within narrow genetic pools, and without the active introduction of new germplasm, genetic variation declines over time, constraining both immediate and long-term genetic progress.
Based on the maximum-likelihood method, the phylogenetic analysis among the 410 cowpea accessions revealed a close genetic relationship between ssp. cylindrica and ssp. unguiculata, whereas ssp. sesquipedalis formed distinct subclusters. This finding is consistent with previous research findings by Wu et al.69. Despite their genetic distinction, both ssp. sesquipedalis and ssp. cylindrica have been traditionally cultivated in Asia for different agricultural purposes26,70. Ssp. cylindrica is primarily used as livestock feed or a cover crop to control weed growth71. The hierarchical clustering categorized the accessions into two main sections: Q1 (green) and Q2 (red), with additional admixed individuals (grey) and an outer group (yellow). Ssp. sesquipedalis accessions were exclusively found in Q2, forming distinct, blue-marked clusters. This exclusivity indicated a clear genetic separation from other subspecies, confirming their genetic separation from other subspecies. In contrast, ssp. cylindrica was found exclusively in Q1, reinforcing its genetic distinctiveness. Ssp. unguiculata exhibited greater genetic diversity, with accessions distributed across both Q1 and Q2.
Geographical clustering was also observed with accessions from the same region often grouped closely. For instance, accessions from Nepal, Pakistan, and Sri Lanka (South Asia) formed close clusters (Cluster 3), as did accessions from Southeast Asia. This suggests that regional breeding practices and environmental factors have influenced genetic variation. Within Q1 and Q2 populations, several small clusters were identified, often characterized by shared ancestry. Additionally, some accessions of ssp. unguiculata and ssp. sesquipedalis exhibited mixed parental ancestry, highlighting the complex genetic dynamics and historical gene flow within the cowpea population.
We proposed the historical migration patterns of cowpeas into Asia based on genetic analysis. The investigation focused on tracing the migration routes of Asian cowpeas introduced from Africa, employing phylogenetic relationships and gene flow analysis conducted with MEGA 11. The results suggested that the flow of genetic material in cowpea germplasm towards Asia and other regions can be traced back to its origins in West and East Africa, corroborating earlier findings by Xiong et al. (2016). Cowpea domestication in Africa predates 1500 BC72,73, with recent findings suggesting a dual center of domestication37. It is suggested that the cowpea was first introduced into Asia around 1500 BC during the Neolithic period, establishing a secondary center of origin in India. As cowpea spread through Asia due to different climatic conditions, the selection for fresh pod consumption led to the emergence of the cultivated group V. unguiculata ssp. unguiculata cv. sesquipedalis in regions like Thailand, China, the Philippines, and India5,37,59,61,74,75. Findings from our study showed that the few accessions contributed from Ethiopia clustered with South Asian accessions in Q2, indicating a similar ancestry and the possible early introduction of East African germplasm into this region. Most likely, cowpea went to South Asia at some point from East Africa, possibly through trade links70. The distribution of cowpea in Asia was mainly from India, South, East, and Southeast Asia, with Japanese accessions primarily from East and Southeast Asia, consistent with previous findings76,77. The phylogenetic tree suggested two major propagation routes into Japan. In southern Japan, the local varieties of cowpea and ssp. sesquipedalis may have been introduced directly from China or via Taiwan, because Yonakuni jima, one of the islands in Japan where the earlier exploration of food legume species was carried out, is separated from Taiwan by only 160 km, as reported by Nakano et al.77. In northern Japan, cowpeas migrated from Southeast Asia. However, documentation of cowpea’s arrival in Southeast Asia is limited. The close clustering of ssp. sesquipedalis accessions from Southeast Asia with those from Japan indicate a shared lineage. Overall, cowpea migration into Asia and Japan likely followed two routes: Africa-India-South Asia-China-East Asia-Japan, and Africa-India-South Asia-China-Southeast Asia-Japan, with subsequent independent movements from East and Southeast Asia to Japan. These findings highlight the significant role of these two cultivation regions in cowpea transmission to Japan.