Partridge L, Deelen J, Slagboom PE. Facing up to the global challenges of ageing. Nature. 2018;561:45–56.
Collaborators GBDD. Global age-sex-specific fertility, mortality, healthy life expectancy (HALE), and population estimates in 204 countries and territories, 1950–2019: a comprehensive demographic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1160–203.
Harper S. Economic and social implications of aging societies. Science. 2014;346:587–91.
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217.
Guerville F, De Souto BP, Ader I, Andrieu S, Casteilla L, Dray C, et al. Revisiting the hallmarks of aging to identify markers of biological age. J Prev Alzheimers Dis. 2020;7:56–64.
Carter CS. A “gut feeling” to create a 10th hallmark of aging. J Gerontol A Biol Sci Med Sci. 2021;76:1891–4.
Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther. 2022;7:135.
de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. 2022;71:1020–32.
O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93.
O’Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–5.
Buford TW. (Dis)trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5:80.
Kundu P, Blacher E, Elinav E, Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017;171:1481–93.
Bradley E, Haran J. The human gut microbiome and aging. Gut Microbes. 2024;16:2359677.
Ghosh TS, Shanahan F, O’Toole PW. The gut microbiome as a modulator of healthy ageing. Nat Rev Gastroenterol Hepatol. 2022;19:565–84.
Walrath T, Dyamenahalli KU, Hulsebus HJ, McCullough RL, Idrovo JP, Boe DM, et al. Age-related changes in intestinal immunity and the microbiome. J Leukoc Biol. 2021;109:1045–61.
Biragyn A, Ferrucci L. Gut dysbiosis: a potential link between increased cancer risk in ageing and inflammaging. Lancet Oncol. 2018;19:e295–304.
Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M, et al. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. Elife. 2017;6:e27014.
Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21:455-466.e4.
Barcena C, Valdes-Mas R, Mayoral P, Garabaya C, Durand S, Rodriguez F, et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat Med. 2019;25:1234–42.
Chen Y, Zhang S, Zeng B, Zhao J, Yang M, Zhang M, et al. Transplant of microbiota from long-living people to mice reduces aging-related indices and transfers beneficial bacteria. Aging (Albany NY). 2020;12:4778–93.
Jing Y, Wang Q, Bai F, Li Z, Li Y, Liu W, et al. Age-related alterations in gut homeostasis are microbiota dependent. NPJ Biofilms Microbiomes. 2025;11:51.
Best L, Dost T, Esser D, Flor S, Gamarra AM, Haase M, et al. Metabolic modelling reveals the aging-associated decline of host-microbiome metabolic interactions in mice. Nat Microbiol. 2025;10:973–91.
Virk B, Correia G, Dixon DP, Feyst I, Jia J, Oberleitner N, et al. Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC Biol. 2012;10:67.
Han B, Sivaramakrishnan P, Lin CJ, Neve IAA, He J, Tay LWR, et al. Microbial genetic composition tunes host longevity. Cell. 2017;169:1249-1262.e13.
Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, et al. Bacterial nitric oxide extends the lifespan of C. elegans. Cell. 2013;152:818–30.
Kim J, Jo Y, Lim G, Ji Y, Roh JH, Kim WG, et al. A microbiota-derived metabolite, 3-phenyllactic acid, prolongs healthspan by enhancing mitochondrial function and stress resilience via SKN-1/ATFS-1 in C. elegans. Nat Commun. 2024;15:10773.
Brummel T, Ching A, Seroude L, Simon AF, Benzer S. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A. 2004;101:12974–9.
Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–52.
Keebaugh ES, Yamada R, Ja WW. The nutritional environment influences the impact of microbes on Drosophila melanogaster life span. MBio. 2019;10:e00885–19.
Obata F, Fons CO, Gould AP. Early-life exposure to low-dose oxidants can increase longevity via microbiome remodelling in Drosophila. Nat Commun. 2018;9:975.
Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–22.
Li H, Qi Y, Jasper H. Preventing age-related decline of gut compartmentalization limits microbiota dysbiosis and extends lifespan. Cell Host Microbe. 2016;19:240–53.
Shukla AK, Johnson K, Giniger E. Common features of aging fail to occur in Drosophila raised without a bacterial microbiome. iScience. 2021;24:102703.
Satokari R, Gronroos T, Laitinen K, Salminen S, Isolauri E. Bifidobacterium and lactobacillus DNA in the human placenta. Lett Appl Microbiol. 2009;48:8–12.
Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra265.
Rautava S, Collado MC, Salminen S, Isolauri E. Probiotics modulate host-microbe interaction in the placenta and fetal gut: a randomized, double-blind, placebo-controlled trial. Neonatology. 2012;102:178–84.
Jimenez E, Fernandez L, Marin ML, Martin R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. 2005;51:270–4.
Younge N, McCann JR, Ballard J, Plunkett C, Akhtar S, Araujo-Perez F, et al. Fetal exposure to the maternal microbiota in humans and mice. JCI Insight. 2019;4:e127806.
Stinson LF, Payne MS, Keelan JA. Planting the seed: origins, composition, and postnatal health significance of the fetal gastrointestinal microbiota. Crit Rev Microbiol. 2017;43:352–69.
de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;572:329–34.
Perez-Munoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5:48.
Kennedy KM, de Goffau MC, Perez-Munoz ME, Arrieta MC, Backhed F, Bork P, et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature. 2023;613:639–49.
Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–5.
Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy. 2008;38:634–42.
Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93.
Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152.
Stokholm J, Thorsen J, Blaser MJ, Rasmussen MA, Hjelmso M, Shah S, et al. Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma. Sci Transl Med. 2020;12:eaax9929.
Hoskinson C, Dai DLY, Del Bel KL, Becker AB, Moraes TJ, Mandhane PJ, et al. Delayed gut microbiota maturation in the first year of life is a hallmark of pediatric allergic disease. Nat Commun. 2023;14:4785.
Martin R, Langa S, Reviriego C, Jiminez E, Marin ML, Xaus J, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143:754–8.
Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69:1–10.
Heikkila MP, Saris PE. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95:471–8.
Collado MC, Delgado S, Maldonado A, Rodriguez JM. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol. 2009;48:523–8.
Hunt KM, Foster JA, Forney LJ, Schutte UM, Beck DL, Abdo Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE. 2011;6:e21313.
Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–51.
Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324-335.e4.
Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–62.
Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185:385–94.
Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884-3898.e11.
Hickman B, Salonen A, Ponsero AJ, Jokela R, Kolho KL, de Vos WM, et al. Gut microbiota wellbeing index predicts overall health in a cohort of 1000 infants. Nat Commun. 2024;15:8323.
Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703.
Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–8.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7.
Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85.
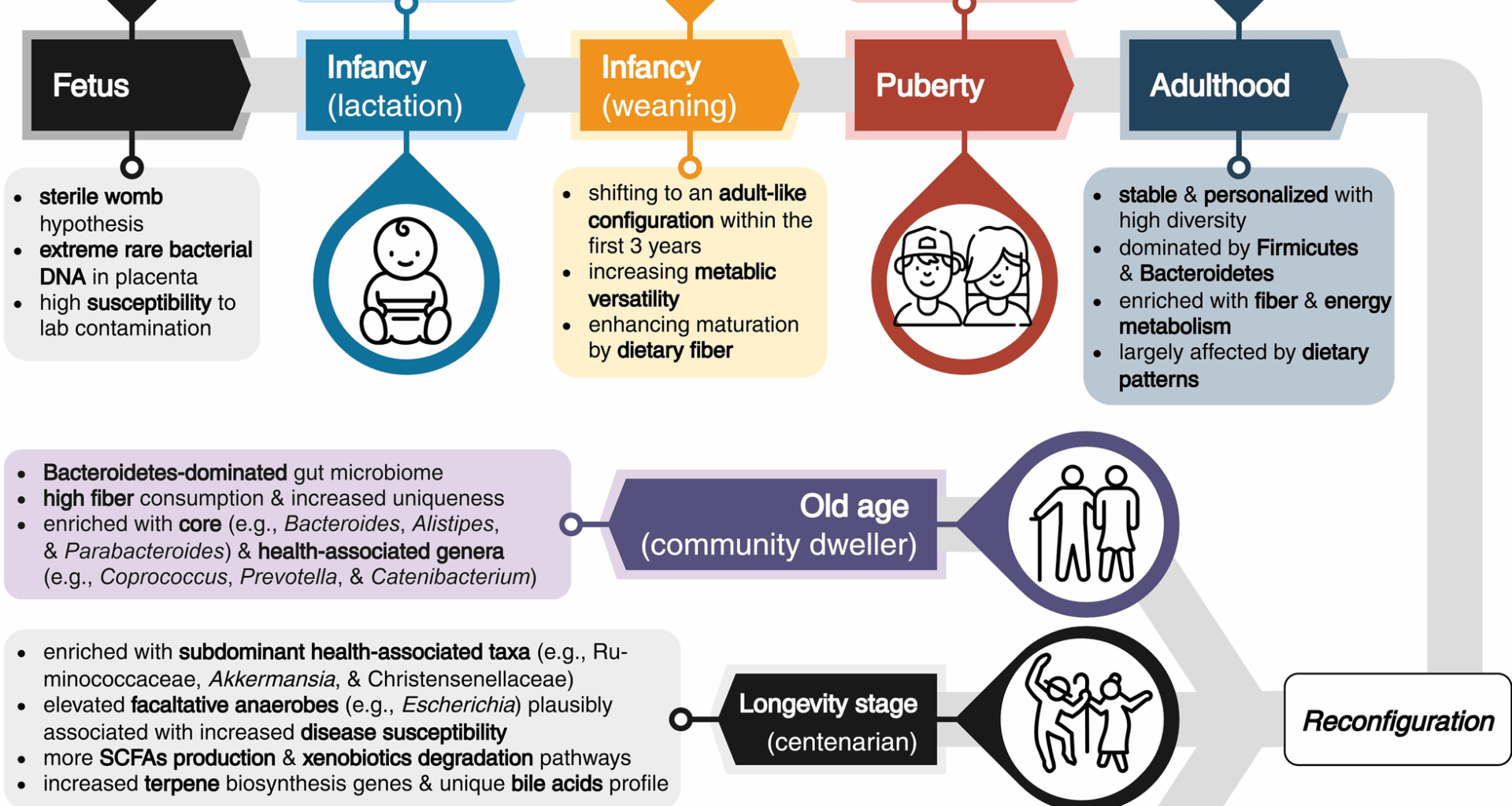
Sawhney SS, Thanert R, Thanert A, Hall-Moore C, Ndao IM, Mahmud B, et al. Gut microbiome evolution from infancy to 8 years of age. Nat Med. 2025. https://doi.org/10.1038/s41591-025-03610-0.
Differding MK, Doyon M, Bouchard L, Perron P, Guerin R, Asselin C, et al. Potential interaction between timing of infant complementary feeding and breastfeeding duration in determination of early childhood gut microbiota composition and BMI. Pediatr Obes. 2020;15:e12642.
Lalli MK, Salo TE, Hakola L, Knip M, Virtanen SM, Vatanen T. Associations between dietary fibers and gut microbiome composition in the EDIA longitudinal infant cohort. Am J Clin Nutr. 2025;121:83–99.
Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ, Paliy O. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77:404–12.
Hollister EB, Riehle K, Luna RA, Weidler EM, Rubio-Gonzales M, Mistretta TA, et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015;3:36.
Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–8.
Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–12.
Weger BD, Gobet C, Yeung J, Martin E, Jimenez S, Betrisey B, et al. The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab. 2019;29:362-382.e8.
Korpela K, Kallio S, Salonen A, Hero M, Kukkonen AK, Miettinen PJ, et al. Gut microbiota develop towards an adult profile in a sex-specific manner during puberty. Sci Rep. 2021;11:23297.
Wang L, Yi Q, Xu H, Liu H, Tan B, Deng H, et al. Alterations in the gut microbiota community are associated with childhood obesity and precocious puberty. BMC Microbiol. 2024;24:311.
Ou Y, Belzer C, Smidt H, de Weerth C. Development of the gut microbiota in the first 14 years of life and its relations to internalizing and externalizing difficulties and social anxiety during puberty. Eur Child Adolesc Psychiatry. 2024;33:847–60.
Murray E, Sharma R, Smith KB, Mar KD, Barve R, Lukasik M, et al. Probiotic consumption during puberty mitigates LPS-induced immune responses and protects against stress-induced depression- and anxiety-like behaviors in adulthood in a sex-specific manner. Brain Behav Immun. 2019;81:198–212.
Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14.
Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–6.
McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, et al. American gut: an open platform for citizen science microbiome research. mSystems. 2018;3:e00031-18.
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–41.
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60.
Lu J, Zhang L, Zhai Q, Zhao J, Zhang H, Lee YK, et al. Chinese gut microbiota and its associations with staple food type, ethnicity, and urbanization. NPJ Biofilms Microbiomes. 2021;7:71.
Tian C, Zhang T, Zhuang D, Luo Y, Li T, Zhao F, et al. Industrialization drives the gut microbiome and resistome of the Chinese populations. mSystems. 2025;10:e0137224.
Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, et al. Microbiome connections with host metabolism and habitual diet from 1098 deeply phenotyped individuals. Nat Med. 2021;27:321–32.
Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–4.
Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–9.
Gacesa R, Kurilshikov A, Vich Vila A, Sinha T, Klaassen MAY, Bolte LA, et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature. 2022;604:732–9.
Nishijima S, Suda W, Oshima K, Kim SW, Hirose Y, Morita H, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23:125–33.
Park J, Kato K, Murakami H, Hosomi K, Tanisawa K, Nakagata T, et al. Comprehensive analysis of gut microbiota of a healthy population and covariates affecting microbial variation in two large Japanese cohorts. BMC Microbiol. 2021;21:151.
Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30.
Abdill RJ, Graham SP, Rubinetti V, Ahmadian M, Hicks P, Chetty A, et al. Integration of 168,000 samples reveals global patterns of the human gut microbiome. Cell. 2025;188:1100-1118.e17.
Rosenberg E. Diversity of bacteria within the human gut and its contribution to the functional unity of holobionts. NPJ Biofilms Microbiomes. 2024;10:134.
Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–8.
Ives AR, Carpenter SR. Stability and diversity of ecosystems. Science. 2007;317:58–62.
Mancabelli L, Milani C, De Biase R, Bocchio F, Fontana F, Lugli GA, et al. Taxonomic and metabolic development of the human gut microbiome across life stages: a worldwide metagenomic investigation. mSystems. 2024;9:e0129423.
Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439.
Safarchi A, Al-Qadami G, Tran CD, Conlon M. Understanding dysbiosis and resilience in the human gut microbiome: biomarkers, interventions, and challenges. Front Microbiol. 2025;16:1559521.
Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343ra381.
Li X, Brejnrod A, Thorsen J, Zachariasen T, Trivedi U, Russel J, et al. Differential responses of the gut microbiome and resistome to antibiotic exposures in infants and adults. Nat Commun. 2023;14:8526.
Zaura E, Brandt BW, Teixeira de Mattos MJ, Buijs MJ, Caspers MP, Rashid MU, et al. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. MBio. 2015;6:e01693-01615.
Hildebrand F, Gossmann TI, Frioux C, Ozkurt E, Myers PN, Ferretti P, et al. Dispersal strategies shape persistence and evolution of human gut bacteria. Cell Host Microbe. 2021;29:1167-1176.e9.
Olsson LM, Boulund F, Nilsson S, Khan MT, Gummesson A, Fagerberg L, et al. Dynamics of the normal gut microbiota: a longitudinal one-year population study in Sweden. Cell Host Microbe. 2022;30:726-739.e3.
Han N, Zhang T, Qiang Y, Peng X, Li X, Zhang W. Time-scale analysis of the long-term variability of human gut microbiota characteristics in Chinese individuals. Commun Biol. 2022;5:1414.
Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–99.
Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191.
Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731–43.
Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, et al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48:1407–12.
Ruhlemann MC, Hermes BM, Bang C, Doms S, Moitinho-Silva L, Thingholm LB, et al. Genome-wide association study in 8956 German individuals identifies influence of ABO histo-blood groups on gut microbiome. Nat Genet. 2021;53:147–55.
Lopera-Maya EA, Kurilshikov A, van der Graaf A, Hu S, Andreu-Sanchez S, Chen L, et al. Effect of host genetics on the gut microbiome in 7738 participants of the Dutch Microbiome Project. Nat Genet. 2022;54:143–51.
Qin Y, Havulinna AS, Liu Y, Jousilahti P, Ritchie SC, Tokolyi A, et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat Genet. 2022;54:134–42.
Wang J, Thingholm LB, Skieceviciene J, Rausch P, Kummen M, Hov JR, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–406.
Liu X, Tang S, Zhong H, Tong X, Jie Z, Ding Q, et al. A genome-wide association study for gut metagenome in Chinese adults illuminates complex diseases. Cell Discov. 2021;7:9.
Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5.
Barker-Tejeda TC, Zubeldia-Varela E, Macias-Camero A, Alonso L, Martin-Antoniano IA, Rey-Stolle MF, et al. Comparative characterization of the infant gut microbiome and their maternal lineage by a multi-omics approach. Nat Commun. 2024;15:3004.
Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, et al. Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE. 2010;5:e10667.
Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, et al. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY). 2013;5:902–12.
Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Sci. 2012;338:120–3.
Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932–42.
Yu LC, Wei SC, Li YH, Lin PY, Chang XY, Weng JP, et al. Invasive pathobionts contribute to colon cancer initiation by counterbalancing epithelial antimicrobial responses. Cell Mol Gastroenterol Hepatol. 2022;13:57–79.
Pai YC, Li YH, Turner JR, Yu LC. Transepithelial barrier dysfunction drives microbiota dysbiosis to initiate epithelial clock-driven inflammation. J Crohns Colitis. 2023;17:1471–88.
Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–91.
Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84.
Jeffery IB, Lynch DB, O’Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016;10:170–82.
Kheirbek RE, Fokar A, Shara N, Bell-Wilson LK, Moore HJ, Olsen E, et al. Characteristics and incidence of chronic illness in community-dwelling predominantly male US veteran centenarians. J Am Geriatr Soc. 2017;65:2100–6.
Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, et al. Gut microbiota and extreme longevity. Curr Biol. 2016;26:1480–5.
Kong F, Hua Y, Zeng B, Ning R, Li Y, Zhao J. Gut microbiota signatures of longevity. Curr Biol. 2016;26:R832–3.
Wang F, Yu T, Huang G, Cai D, Liang X, Su H, et al. Gut microbiota community and its assembly associated with age and diet in Chinese centenarians. J Microbiol Biotechnol. 2015;25:1195–204.
Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90.
Santoro A, Ostan R, Candela M, Biagi E, Brigidi P, Capri M, et al. Gut microbiota changes in the extreme decades of human life: a focus on centenarians. Cell Mol Life Sci. 2018;75:129–48.
Pang S, Chen X, Lu Z, Meng L, Huang Y, Yu X, et al. Longevity of centenarians is reflected by the gut microbiome with youth-associated signatures. Nat Aging. 2023;3:436–49.
Wilmanski T, Diener C, Rappaport N, Patwardhan S, Wiedrick J, Lapidus J, et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat Metab. 2021;3:274–86.
Ghosh TS, Shanahan F, O’Toole PW. Toward an improved definition of a healthy microbiome for healthy aging. Nat Aging. 2022;2:1054–69.
Cheng S, Larson MG, McCabe EL, Murabito JM, Rhee EP, Ho JE, et al. Distinct metabolomic signatures are associated with longevity in humans. Nat Commun. 2015;6(1):6791.
Wu L, Zeng T, Zinellu A, Rubino S, Kelvin DJ, Carru C. A cross-sectional study of compositional and functional profiles of gut microbiota in Sardinian centenarians. mSystems. 2019;4:e00325–19.
Wu L, Xie X, Li Y, Liang T, Zhong H, Yang L, et al. Gut microbiota as an antioxidant system in centenarians associated with high antioxidant activities of gut-resident Lactobacillus. NPJ Biofilms Microbiomes. 2022;8:102.
Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature. 2021;599:458–64.
Liu S, Zhang Z, Wang X, Ma Y, Ruan H, Wu X, et al. Biosynthetic potential of the gut microbiome in longevous populations. Gut Microbes. 2024;16:2426623.
Si J, Vazquez-Castellanos JF, Gregory AC, Decommer L, Rymenans L, Proost S, et al. Long-term life history predicts current gut microbiome in a population-based cohort study. Nat Aging. 2022;2:885–95.
Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4-9.
Nikolich-Zugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat Immunol. 2018;19:10–9.
Bosco N, Noti M. The aging gut microbiome and its impact on host immunity. Genes Immun. 2021;22:289–303.
Correa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. 2016;5:e73.
Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8.
Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6.
Cryan JF, O’Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, et al. The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013.
Solanki R, Karande A, Ranganathan P. Emerging role of gut microbiota dysbiosis in neuroinflammation and neurodegeneration. Front Neurol. 2023;14:1149618.
Rothhammer V, Borucki DM, Tjon EC, Takenaka MC, Chao CC, Ardura-Fabregat A, et al. Microglial control of astrocytes in response to microbial metabolites. Nature. 2018;557:724–8.
Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–77.
Erny D, Dokalis N, Mezo C, Castoldi A, Mossad O, Staszewski O, et al. Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metab. 2021;33:2260-2276.e7.
Duscha A, Gisevius B, Hirschberg S, Yissachar N, Stangl GI, Dawin E, et al. Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell. 2020;180:1067-1080.e6.
Liu X, Li X, Xia B, Jin X, Zou Q, Zeng Z, et al. High-fiber diet mitigates maternal obesity-induced cognitive and social dysfunction in the offspring via gut-brain axis. Cell Metab. 2021;33:923-938.e6.
Seo DO, O’Donnell D, Jain N, Ulrich JD, Herz J, Li Y, et al. ApoE isoform- and microbiota-dependent progression of neurodegeneration in a mouse model of tauopathy. Science. 2023;379:eadd1236.
Colombo AV, Sadler RK, Llovera G, Singh V, Roth S, Heindl S, et al. Microbiota-derived short chain fatty acids modulate microglia and promote Abeta plaque deposition. Elife. 2021;10:e59826.
Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167(1469–1480):e1412.
Shin C, Lim Y, Lim H, Ahn TB. Plasma short-chain fatty acids in patients with Parkinson’s disease. Mov Disord. 2020;35:1021–7.
Yang X, Ai P, He X, Mo C, Zhang Y, Xu S, et al. Parkinson’s disease is associated with impaired gut-blood barrier for short-chain fatty acids. Mov Disord. 2022;37:1634–43.
Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, et al. Signatures of early frailty in the gut microbiota. Genome Med. 2016;8:8.
Almeida HM, Sardeli AV, Conway J, Duggal NA, Cavaglieri CR. Comparison between frail and non-frail older adults’ gut microbiota: a systematic review and meta-analysis. Ageing Res Rev. 2022;82:101773.
Lahiri S, Kim H, Garcia-Perez I, Reza MM, Martin KA, Kundu P, et al. The gut microbiota influences skeletal muscle mass and function in mice. Sci Transl Med. 2019;11:eaan5662.
Li G, Jin B, Fan Z. Mechanisms involved in gut microbiota regulation of skeletal muscle. Oxid Med Cell Longev. 2022;2022:2151191.
Han DS, Wu WK, Liu PY, Yang YT, Hsu HC, Kuo CH, et al. Differences in the gut microbiome and reduced fecal butyrate in elders with low skeletal muscle mass. Clin Nutr. 2022;41:1491–500.
Otsuka R, Zhang S, Furuya K, Tange C, Sala G, Ando F, et al. Association between short-chain fatty acid intake and development of muscle strength loss among community-dwelling older Japanese adults. Exp Gerontol. 2023;173:112080.
Liu C, Wong PY, Wang Q, Wong HY, Huang T, Cui C, et al. Short-chain fatty acids enhance muscle mass and function through the activation of mTOR signalling pathways in sarcopenic mice. J Cachexia Sarcopenia Muscle. 2024;15:2387–401.
Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8.
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63.
Fackelmann G, Manghi P, Carlino N, Heidrich V, Piccinno G, Ricci L, et al. Gut microbiome signatures of vegan, vegetarian and omnivore diets and associated health outcomes across 21,561 individuals. Nat Microbiol. 2025;10:41–52.
De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–21.
Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri M, et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut. 2020;69:1218–28.
Marseglia A, Xu W, Fratiglioni L, Fabbri C, Berendsen AAM, Bialecka-Debek A, et al. Effect of the NU-AGE diet on cognitive functioning in older adults: a randomized controlled trial. Front Physiol. 2018;9:349.
Xu Z, Knight R. Dietary effects on human gut microbiome diversity. Br J Nutr. 2015;113(Suppl):S1-5.
Tessier AJ, Wang F, Korat AA, Eliassen AH, Chavarro J, Grodstein F, et al. Optimal dietary patterns for healthy aging. Nat Med. 2025. https://doi.org/10.1038/s41591-025-03570-5.
Di Francesco A, Deighan AG, Litichevskiy L, Chen Z, Luciano A, Robinson L, et al. Dietary restriction impacts health and lifespan of genetically diverse mice. Nature. 2024;634:684–92.
Litichevskiy L, Considine M, Gill J, Shandar V, Cox TO, Descamps HC, et al. Gut metagenomes reveal interactions between dietary restriction, ageing and the microbiome in genetically diverse mice. Nat Microbiol. 2025. https://doi.org/10.1038/s41564-025-01963-3.
Ruiz A, Cerdo T, Jauregui R, Pieper DH, Marcos A, Clemente A, et al. One-year calorie restriction impacts gut microbial composition but not its metabolic performance in obese adolescents. Environ Microbiol. 2017;19:1536–51.
Pisanu S, Palmas V, Madau V, Casula E, Deledda A, Cusano R, et al. Impact of a moderately hypocaloric mediterranean diet on the gut microbiota composition of Italian obese patients. Nutrients. 2020;12:2707.
Flanagan EW, Most J, Mey JT, Redman LM. Calorie restriction and aging in humans. Annu Rev Nutr. 2020;40:105–33.
Severinsen MCK, Pedersen BK. Muscle-organ crosstalk: the emerging roles of myokines. Endocr Rev. 2020;41:594–609.
Qiu Y, Fernandez-Garcia B, Lehmann HI, Li G, Kroemer G, Lopez-Otin C, et al. Exercise sustains the hallmarks of health. J Sport Health Sci. 2023;12:8–35.
Mailing LJ, Allen JM, Buford TW, Fields CJ, Woods JA. Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc Sport Sci Rev. 2019;47:75–85.
Hawley JA, Forster SC, Giles EM. Exercise, the gut microbiome and gastrointestinal diseases: therapeutic impact and molecular mechanisms. Gastroenterology. 2025. https://doi.org/10.1053/j.gastro.2025.01.224.
Varghese S, Rao S, Khattak A, Zamir F, Chaari A. Physical exercise and the gut microbiome: a bidirectional relationship influencing health and performance. Nutrients. 2024;16:3663.
Lai ZL, Tseng CH, Ho HJ, Cheung CKY, Lin JY, Chen YJ, et al. Fecal microbiota transplantation confers beneficial metabolic effects of diet and exercise on diet-induced obese mice. Sci Rep. 2018;8:15625.
Clarke SF, Murphy EF, O’Sullivan O, Lucey AJ, Humphreys M, Hogan A, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–20.
Barton W, Penney NC, Cronin O, Garcia-Perez I, Molloy MG, Holmes E, et al. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. 2018;67:625–33.
Bressa C, Bailen-Andrino M, Perez-Santiago J, Gonzalez-Soltero R, Perez M, Montalvo-Lominchar MG, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE. 2017;12:e0171352.
Munukka E, Ahtiainen JP, Puigbo P, Jalkanen S, Pahkala K, Keskitalo A, et al. Six-week endurance exercise alters gut metagenome that is not reflected in systemic metabolism in over-weight women. Front Microbiol. 2018;9:2323.
Martin D, Bonneau M, Orfila L, Horeau M, Hazon M, Demay R, et al. Atypical gut microbial ecosystem from athletes with very high exercise capacity improves insulin sensitivity and muscle glycogen store in mice. Cell Rep. 2025;44:115448.
Erlandson KM, Liu J, Johnson R, Dillon S, Jankowski CM, Kroehl M, et al. An exercise intervention alters stool microbiota and metabolites among older, sedentary adults. Ther Adv Infect Dis. 2021;8:20499361211027067.
Zhu Q, Jiang S, Du G. Effects of exercise frequency on the gut microbiota in elderly individuals. Microbiol Open. 2020;9:e1053.
Clauss M, Gerard P, Mosca A, Leclerc M. Interplay between exercise and gut microbiome in the context of human health and performance. Front Nutr. 2021;8:637010.
van Wijck K, Lenaerts K, van Loon LJ, Peters WH, Buurman WA, Dejong CH. Exercise-induced splanchnic hypoperfusion results in gut dysfunction in healthy men. PLoS ONE. 2011;6:e22366.
Karl JP, Margolis LM, Madslien EH, Murphy NE, Castellani JW, Gundersen Y, et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am J Physiol Gastrointest Liver Physiol. 2017;312:G559–71.
Jayanama K, Theou O. Effects of probiotics and prebiotics on frailty and ageing: a narrative review. Curr Clin Pharmacol. 2020;15:183–92.
Ale EC, Binetti AG. Role of probiotics, prebiotics, and synbiotics in the elderly: insights into their applications. Front Microbiol. 2021;12:631254.
Hong CT, Chen JH, Huang TW. Probiotics treatment for Parkinson disease: a systematic review and meta-analysis of clinical trials. Aging (Albany NY). 2022;14:7014–25.
Shokri-Mashhadi N, Navab F, Ansari S, Rouhani MH, Hajhashemy Z, Saraf-Bank S. A meta-analysis of the effect of probiotic administration on age-related sarcopenia. Food Sci Nutr. 2023;11:4975–87.
Setbo E, Campbell K, O’Cuiv P, Hubbard R. Utility of probiotics for maintenance or improvement of health status in older people—a scoping review. J Nutr Health Aging. 2019;23:364–72.
Jukic Peladic N, Dell’Aquila G, Carrieri B, Maggio M, Cherubini A, Orlandoni P. Potential role of probiotics for inflammaging: a narrative review. Nutrients. 2021;13:2919.
Recharla N, Choi J, Puligundla P, Park SJ, Lee HJ. Impact of probiotics on cognition and constipation in the elderly: a meta-analysis. Heliyon. 2023;9:e18306.
Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388-1405.e1.
Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. 2021;18:649–67.
Arnold JW, Roach J, Fabela S, Moorfield E, Ding S, Blue E, et al. The pleiotropic effects of prebiotic galacto-oligosaccharides on the aging gut. Microbiome. 2021;9:31.
Kadyan S, Park G, Singh P, Arjmandi B, Nagpal R. Prebiotic mechanisms of resistant starches from dietary beans and pulses on gut microbiome and metabolic health in a humanized murine model of aging. Front Nutr. 2023;10:1106463.
Kadyan S, Park G, Wang B, Nagpal R. Dietary fiber modulates gut microbiome and metabolome in a host sex-specific manner in a murine model of aging. Front Mol Biosci. 2023;10:1182643.
Lee SH, You HS, Kang HG, Kang SS, Hyun SH. Association between altered blood parameters and gut microbiota after synbiotic intake in healthy, elderly Korean women. Nutrients. 2020;12:3112.
Wang S, Ahmadi S, Nagpal R, Jain S, Mishra SP, Kavanagh K, et al. Lipoteichoic acid from the cell wall of a heat killed Lactobacillus paracasei D3–5 ameliorates aging-related leaky gut, inflammation and improves physical and cognitive functions: from C. elegans to mice. Geroscience. 2020;42:333–52.
Powell DN, Swimm A, Sonowal R, Bretin A, Gewirtz AT, Jones RM, et al. Indoles from the commensal microbiota act via the AHR and IL-10 to tune the cellular composition of the colonic epithelium during aging. Proc Natl Acad Sci U S A. 2020;117:21519–26.
Chen P, Chen F, Lei J, Zhou B. Gut microbial metabolite urolithin B attenuates intestinal immunity function in vivo in aging mice and in vitro in HT29 cells by regulating oxidative stress and inflammatory signalling. Food Funct. 2021;12:11938–55.
Kolonics A, Bori Z, Torma F, Abraham D, Feher J, Radak Z. Exercise combined with postbiotics treatment results in synergistic improvement of mitochondrial function in the brain of male transgenic mice for Alzheimer’s disease. BMC Neurosci. 2023;24:68.
El Haddad L, Mendoza JF, Jobin C. Bacteriophage-mediated manipulations of microbiota in gastrointestinal diseases. Front Microbiol. 2022;13:1055427.
Fujiki J, Schnabl B. Phage therapy: targeting intestinal bacterial microbiota for the treatment of liver diseases. JHEP Rep. 2023;5:100909.
Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575:505–11.
Federici S, Kredo-Russo S, Valdes-Mas R, Kviatcovsky D, Weinstock E, Matiuhin Y, et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell. 2022;185:2879-2898.e4.
Ichikawa M, Nakamoto N, Kredo-Russo S, Weinstock E, Weiner IN, Khabra E, et al. Bacteriophage therapy against pathological Klebsiella pneumoniae ameliorates the course of primary sclerosing cholangitis. Nat Commun. 2023;14:3261.
Gan L, Feng Y, Du B, Fu H, Tian Z, Xue G, et al. Bacteriophage targeting microbiota alleviates non-alcoholic fatty liver disease induced by high alcohol-producing Klebsiella pneumoniae. Nat Commun. 2023;14:3215.
Gindin M, Febvre HP, Rao S, Wallace TC, Weir TL. Bacteriophage for Gastrointestinal Health (PHAGE) study: evaluating the safety and tolerability of supplemental bacteriophage consumption. J Am Coll Nutr. 2019;38:68–75.
Grubb DS, Wrigley SD, Freedman KE, Wei Y, Vazquez AR, Trotter RE, et al. Phage-2 study: supplemental bacteriophages extend Bifidobacterium animalis subsp. lactis BL04 benefits on gut health and microbiota in healthy adults. Nutrients. 2020;12(Suppl):2474.
Shen X, Wang C, Zhou X, Zhou W, Hornburg D, Wu S, et al. Nonlinear dynamics of multi-omics profiles during human aging. Nat Aging. 2024;4:1619–34.
Yang H, Wang T, Qian C, Wang H, Yu D, Shi M, et al. Gut microbial-derived phenylacetylglutamine accelerates host cellular senescence. Nat Aging. 2025;5:401–18.
Tseng CH, Wong S, Yu J, Lee YY, Terauchi J, Lai HC, et al. Development of live biotherapeutic products: a position statement of Asia-Pacific Microbiota Consortium. Gut. 2025;74:706–13.