Monod, J. Chance and Necessity: an Essay on the Natural Philosophy of Modern Biology (Vintage Books, 1972).
Blount, Z. D., Lenski, R. E. & Losos, J. B. Contingency and determinism in evolution: replaying life’s tape. Science 362, eaam5979 (2018).
Luria, S. E. & Delbrück, M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511 (1943). This foundational paper introduced the fluctuation test, providing the first experimental evidence that bacterial mutations arise randomly rather than in response to selective pressure.
Lederberg, J. & Lederberg, E. M. Replica plating and indirect selection of bacterial mutants. J. Bacteriol. 63, 399–406 (1952).
Helling, R. B., Vargas, C. N. & Adams, J. Evolution of Escherichia coli during growth in a constant environment. Genetics 116, 349–358 (1987).
Lenski, R. E., Rose, M. R., Simpson, S. C. & Tadler, S. C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341 (1991). This landmark paper reported the first results from the E. coli LTEE, now the longest-running evolution experiment, which has transformed our understanding of bacterial adaptation.
Zamenhof, S. & Eichhorn, H. H. Study of microbial evolution through loss of biosynthetic functions: establishment of “defective” mutants. Nature 216, 456–458 (1967).
Lenski, R. E. & Levin, B. R. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 125, 585–602 (1985).
Tenaillon, O. et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536, 165–170 (2016). This study sequenced hundreds of clones isolated from the E. coli LTEE, revealing widespread parallelism in the targets of mutations across replicate populations.
Chou, H.-H., Chiu, H.-C., Delaney, N. F., Segrè, D. & Marx, C. J. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332, 1190–1192 (2011).
Khan, A. I., Dinh, D. M., Schneider, D., Lenski, R. E. & Cooper, T. F. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332, 1193–1196 (2011).
Kryazhimskiy, S., Rice, D. P., Jerison, E. R. & Desai, M. M. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344, 1519–1522 (2014).
Rozen, D. E. & Lenski, R. E. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am. Nat. 155, 24–35 (2000). This study uncovered the spontaneous emergence of a simple two-ecotype ecosystem in the E. coli LTEE, driven by metabolic cross-feeding, which has persisted for tens of thousands of generations.
Rozen, D. E., Philippe, N., Arjan de Visser, J., Lenski, R. E. & Schneider, D. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol. Lett. 12, 34–44 (2009).
Herron, M. D. & Doebeli, M. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 11, e1001490 (2013).
Barrick, J. E. et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247 (2009).
Miller, C. R., Joyce, P. & Wichman, H. A. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187, 185 (2011).
Levin, B. R., Stewart, F. M. & Chao, L. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111, 3–24 (1977).
Hegreness, M., Shoresh, N., Hartl, D. & Kishony, R. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311, 1615–1617 (2006).
Jagdish, T. & Nguyen Ba, A. N. Microbial experimental evolution in a massively multiplexed and high-throughput era. Curr. Opin. Genet. Dev. 75, 101943 (2022).
Desai, M. M. Statistical questions in experimental evolution. J. Stat. Mech. Theory Exp. 2013, P01003 (2013).
Venkataram, S. et al. Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell 166, 1585–1596.e22 (2016).
Tenaillon, O. et al. The molecular diversity of adaptive convergence. Science 335, 457–461 (2012).
Jerison, E. R., Nguyen Ba, A. N., Desai, M. M. & Kryazhimskiy, S. Chance and necessity in the pleiotropic consequences of adaptation for budding yeast. Nat. Ecol. Evol. 4, 601–611 (2020).
Lang, G. I. et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500, 571–574 (2013).
Good, B. H., McDonald, M. J., Barrick, J. E., Lenski, R. E. & Desai, M. M. The dynamics of molecular evolution over 60,000 generations. Nature 551, 45–50 (2017). This study performed time-resolved metagenomic sequencing on all 12 LTEE populations over 60,000 generations of evolution, revealing that most (9/12) populations showed signatures of multiple ecotypes coexisting over tens of thousands of generations.
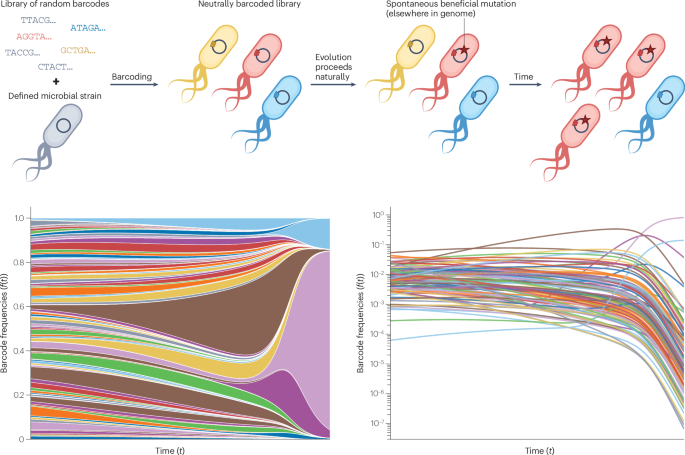
Levy, S. F. et al. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature 519, 181–186 (2015). This study introduced high-resolution lineage tracking using DNA barcoding, revolutionizing our ability to observe evolutionary dynamics in real time.
Wetmore, K. M. et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6, e00306–e00315 (2015).
Nguyen Ba, A. N. et al. High-resolution lineage tracking reveals travelling wave of adaptation in laboratory yeast. Nature 575, 494–499 (2019). This work demonstrated the power of re-barcoding to maintain barcode diversity during evolution, allowing for the high-resolution observation of evolutionary dynamics beyond the first fixation event.
Price, M. N. et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503–509 (2018).
Ang, R. M. L., Chen, S.-A. A., Kern, A. F., Xie, Y. & Fraser, H. B. Widespread epistasis among beneficial genetic variants revealed by high-throughput genome editing. Cell Genomics 3, 100260 (2023).
Dadonaite, B. et al. Spike deep mutational scanning helps predict success of SARS-CoV-2 clades. Nature 631, 617–626 (2024).
Li, F., Salit, M. L. & Levy, S. F. Unbiased fitness estimation of pooled barcode or amplicon sequencing studies. Cell Syst. 7, 521–525.e4 (2018).
Limdi, A. & Baym, M. Resolving deleterious and near-neutral effects requires different pooled fitness assay designs. J. Mol. Evol. 91, 325–333 (2023).
Jasinska, W. et al. Chromosomal barcoding of E. coli populations reveals lineage diversity dynamics at high resolution. Nat. Ecol. Evol. 4, 437–452 (2020).
Good, B. H., Rouzine, I. M., Balick, D. J., Hallatschek, O. & Desai, M. M. Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proc. Natl Acad. Sci. USA 109, 4950–4955 (2012).
Smith, A. M. et al. Quantitative phenotyping via deep barcode sequencing. Genome Res. 19, 1836–1842 (2009).
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002).
Ascensao, J. A., Wetmore, K. M., Good, B. H., Arkin, A. P. & Hallatschek, O. Quantifying the local adaptive landscape of a nascent bacterial community. Nat. Commun. 14, 248 (2023). This paper used a barcoded knockout approach to uncover how the ecological interactions between E. coli LTEE ecotypes impact evolutionary adaptability, revealing that community composition and other ecological conditions pervasively and predictably impact knockout fitness effects.
Couce, A. et al. Changing fitness effects of mutations through long-term bacterial evolution. Science 383, eadd1417 (2024). This study used a barcoded knockout approach to quantify how the distribution of fitness effects changed over time in the E. coli LTEE, revealing strong diminishing returns epistasis and shifting knockout fitness effects.
Loes, A. N. et al. High-throughput sequencing-based neutralization assay reveals how repeated vaccinations impact titers to recent human H1N1 influenza strains. J. Virol. 98, e00689–24 (2024).
Mutalik, V. K. et al. Dual-barcoded shotgun expression library sequencing for high-throughput characterization of functional traits in bacteria. Nat. Commun. 10, 308 (2019).
Johnson, M. S., Martsul, A., Kryazhimskiy, S. & Desai, M. M. Higher-fitness yeast genotypes are less robust to deleterious mutations. Science 366, 490–493 (2019).
Peters, J. M. et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat. Microbiol. 4, 244–250 (2019).
Hawkins, J. S. et al. Mismatch-CRISPRi reveals the co-varying expression–fitness relationships of essential genes in Escherichia coli and Bacillus subtilis. Cell Syst. 11, 523–535.e9 (2020).
Sharon, E. et al. Functional genetic variants revealed by massively parallel precise genome editing. Cell 175, 544–557.e16 (2018).
Chen, S.-A. A., Kern, A. F., Ang, R. M. L., Xie, Y. & Fraser, H. B. Gene-by-environment interactions are pervasive among natural genetic variants. Cell Genomics 3, 100273 (2023).
Lee, J. M. et al. Deep mutational scanning of hemagglutinin helps predict evolutionary fates of human H3N2 influenza variants. Proc. Natl. Acad. Sci. 115, E8276–E8285 (2018).
Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e20 (2020).
Bakerlee, C. W., Nguyen Ba, A. N., Shulgina, Y., Rojas Echenique, J. I. & Desai, M. M. Idiosyncratic epistasis leads to global fitness-correlated trends. Science 376, 630–635 (2022).
Wu, N. C., Dai, L., Olson, C. A., Lloyd-Smith, J. O. & Sun, R. Adaptation in protein fitness landscapes is facilitated by indirect paths. eLife 5, e16965 (2016).
Pokusaeva, V. O. et al. An experimental assay of the interactions of amino acids from orthologous sequences shaping a complex fitness landscape. PLoS Genet. 15, e1008079 (2019).
Faure, A. J. et al. Mapping the energetic and allosteric landscapes of protein binding domains. Nature 604, 175–183 (2022).
Kuo, S.-T. et al. Global fitness landscapes of the Shine–Dalgarno sequence. Genome Res. 30, 711–723 (2020).
Domingo, J., Diss, G. & Lehner, B. Pairwise and higher-order genetic interactions during the evolution of a tRNA. Nature 558, 117–121 (2018).
Weinreich, D. M., Delaney, N. F., Depristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006).
Papkou, A., Garcia-Pastor, L., Escudero, J. A. & Wagner, A. A rugged yet easily navigable fitness landscape. Science 382, eadh3860 (2023).
Phillips, A. M. et al. Binding affinity landscapes constrain the evolution of broadly neutralizing anti-influenza antibodies. eLife 10, e71393 (2021).
Phillips, A. M. et al. Hierarchical sequence-affinity landscapes shape the evolution of breadth in an anti-influenza receptor binding site antibody. eLife 12, e83628 (2023).
Moulana, A. et al. Compensatory epistasis maintains ACE2 affinity in SARS-CoV-2 Omicron BA.1. Nat. Commun. 13, 7011 (2022).
Johnston, K. E. et al. A combinatorially complete epistatic fitness landscape in an enzyme active site. Proc. Natl Acad. Sci. USA 121, e2400439121 (2024).
Achaz, G., Rodriguez-Verdugo, A., Gaut, B. S. & Tenaillon, O. The reproducibility of adaptation in the light of experimental evolution with whole genome sequencing. Adv. Exp. Med. Biol. 781, 211–231 (2014).
Bergelson, J., Kreitman, M., Petrov, D. A., Sanchez, A. & Tikhonov, M. Functional biology in its natural context: a search for emergent simplicity. eLife 10, e67646 (2021).
Lässig, M., Mustonen, V. & Walczak, A. M. Predicting evolution. Nat. Ecol. Evol. 1, 0077 (2017).
Wright, S. Evolution in mendelian populations. Genetics 16, 97–159 (1931).
Fisher, R. The Genetical Theory of Natural Selection (1930).
Wiser, M. J., Ribeck, N. & Lenski, R. E. Long-term dynamics of adaptation in asexual populations. Science 342, 1364–1367 (2013). This paper provided the first comprehensive analysis of fitness trajectories over 50,000 generations of the LTEE, revealing that fitness gains are largely parallel across replicate populations.
Láruson, Á. J., Yeaman, S. & Lotterhos, K. E. The importance of genetic redundancy in evolution. Trends Ecol. Evol. 35, 809–822 (2020).
Favate, J. S., Liang, S., Cope, A. L., Yadavalli, S. S. & Shah, P. The landscape of transcriptional and translational changes over 22 years of bacterial adaptation. eLife 11, e81979 (2022). This study used high-throughput sequencing to measure changes in transcription and translation in E. coli LTEE populations, revealing widespread parallelism across populations.
Grant, N. A., Abdel Magid, A., Franklin, J., Dufour, Y. & Lenski, R. E. Changes in cell size and shape during 50,000 generations of experimental evolution with Escherichia coli. J. Bacteriol. 203, e00469–20 (2021).
Card, K. J., Thomas, M. D., Graves, J. L., Barrick, J. E. & Lenski, R. E. Genomic evolution of antibiotic resistance is contingent on genetic background following a long-term experiment with Escherichia coli. Proc. Natl Acad. Sci. USA 118, e2016886118 (2021).
Favate, J. S. et al. Linking genotypic and phenotypic changes in the E. coli long-term evolution experiment using metabolomics. eLife 12, RP87039 (2023).
Lukačišinová, M., Fernando, B. & Bollenbach, T. Highly parallel lab evolution reveals that epistasis can curb the evolution of antibiotic resistance. Nat. Commun. 11, 3105 (2020).
Reddy, G. & Desai, M. M. Global epistasis emerges from a generic model of a complex trait. eLife 10, e64740 (2021). This theoretical work developed a simple statistical model showing how genetic variants on the same background interact to influence trait values, demonstrating that diminishing returns and increasing costs epistasis naturally emerge from this model.
Blount, Z. D., Borland, C. Z. & Lenski, R. E. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 7899–7906 (2008). This landmark study documented the evolution of citrate use in the E. coli LTEE, demonstrating how historical contingency can influence the emergence of evolutionary innovations.
Frenkel, E. M. et al. Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations. Proc. Natl Acad. Sci. USA 112, 11306–11311 (2015).
Kinnersley, M. A., Holben, W. E. & Rosenzweig, F. E unibus plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet. 5, e1000713 (2009).
D’Souza, G. & Kost, C. Experimental evolution of metabolic dependency in bacteria. PLoS Genet. 12, e1006364 (2016).
Ascensao, J. A. et al. Rediversification following ecotype isolation reveals hidden adaptive potential. Curr. Biol. 34, 855–867.e6 (2024).
Rainey, P. B. & Travisano, M. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72 (1998).
Flohr, R. C. E., Blom, C. J., Rainey, Paul, B. & Beaumont, H. J. E. Founder niche constrains evolutionary adaptive radiation. Proc. Natl Acad. Sci. USA 110, 20663–20668 (2013).
Lenski, R. E. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J. 11, 2181–2194 (2017).
Johnson, M. S. et al. Phenotypic and molecular evolution across 10,000 generations in laboratory budding yeast populations. eLife 10, e63910 (2021).
Turner, C. B., Blount, Z. D. & Lenski, R. E. Replaying evolution to test the cause of extinction of one ecotype in an experimentally evolved population. PLoS ONE 10, e0142050 (2015).
Großkopf, T. et al. Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment. BMC Evol. Biol. 16, 163 (2016).
Rozen, D. E., Schneider, D. & Lenski, R. E. Long-term experimental evolution in Escherichia coli. XIII. Phylogenetic history of a balanced polymorphism. J. Mol. Evol. 61, 171–180 (2005).
Le Gac, M., Plucain, J., Hindré, T., Lenski, R. E. & Schneider, D. Ecological and evolutionary dynamics of coexisting lineages during a long-term experiment with Escherichia coli. Proc. Natl Acad. Sci. USA 109, 9487–9492 (2012).
Plucain, J. et al. Epistasis and allele specificity in the emergence of a stable polymorphism in Escherichia coli. Science 343, 1366–1369 (2014).
Maddamsetti, R., Lenski, R. E. & Barrick, J. E. Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics 200, 619–631 (2015).
Zhao, S. et al. Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe 25, 656–667.e8 (2019).
Sousa, A. et al. Recurrent reverse evolution maintains polymorphism after strong bottlenecks in commensal gut bacteria. Mol. Biol. Evol. 34, 2879–2892 (2017).
Friedman, J., Higgins, L. M. & Gore, J. Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 1, 0109 (2017).
Goldford, J. E. et al. Emergent simplicity in microbial community assembly. Science 361, 469–474 (2018).
Louca, S. et al. High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol. 1, 0015 (2016).
Louca, S. et al. Functional structure of the bromeliad tank microbiome is strongly shaped by local geochemical conditions. Environ. Microbiol. 19, 3132–3151 (2017).
Marsland, R. et al. Available energy fluxes drive a transition in the diversity, stability, and functional structure of microbial communities. PLoS Comput. Biol. 15, e1006793 (2019).
Ruiz, J. et al. Predictability of the community‐function landscape in wine yeast ecosystems. Mol. Syst. Biol. 19, e11613 (2023).
Diaz-Colunga, J., Skwara, A., Vila, J. C. C., Bajic, D. & Sanchez, A. Global epistasis and the emergence of function in microbial consortia. Cell 187, 3108–3119.e30 (2024).
Lopes, W., Amor, D. R. & Gore, J. Cooperative growth in microbial communities is a driver of multistability. Nat. Commun. 15, 4709 (2024).
Hu, J., Amor, D. R., Barbier, M., Bunin, G. & Gore, J. Emergent phases of ecological diversity and dynamics mapped in microcosms. Science 378, 85–89 (2022).
Lee, H., Bloxham, B. & Gore, J. Resource competition can explain simplicity in microbial community assembly. Proc. Natl Acad. Sci. USA 120, e2212113120 (2023).
Meroz, N., Tovi, N., Sorokin, Y. & Friedman, J. Community composition of microbial microcosms follows simple assembly rules at evolutionary timescales. Nat. Commun. 12, 2891 (2021).
Meroz, N. et al. Evolution in microbial microcosms is highly parallel, regardless of the presence of interacting species. Cell Syst. 15, 930–940.e5 (2024).
Barrick, J. E. & Lenski, R. E. Genome dynamics during experimental evolution. Nat. Rev. Genet. 14, 827–839 (2013).
Gerrish, P. & Lenski, R. The fate of competing beneficial mutations in an asexual population. Genetica 102, 127–144 (1998).
Miralles, R., Gerrish, P. J., Moya, A. & Elena, S. F. Clonal interference and the evolution of RNA viruses. Science 285, 1745–1747 (1999).
Pepin, K. M. & Wichman, H. A. Experimental evolution and genome sequencing reveal variation in levels of clonal interference in large populations of bacteriophage phiX174. BMC Evol. Biol. 8, 85 (2008).
Kvitek, D. J. & Sherlock, G. Whole genome, whole population sequencing reveals that loss of signaling networks is the major adaptive strategy in a constant environment. PLoS Genet. 9, e1003972 (2013).
Desai, M. M., Fisher, D. S. & Murray, A. W. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17, 385–394 (2007).
Wilson, B. A., Garud, N. R., Feder, A. F., Assaf, Z. J. & Pennings, P. S. The population genetics of drug resistance evolution in natural populations of viral, bacterial and eukaryotic pathogens. Mol. Ecol. 25, 42–66 (2016).
Eldholm, V. et al. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol. 15, 490 (2014).
Strelkowa, N. & Lässig, M. Clonal interference in the evolution of influenza. Genetics 192, 671–682 (2012).
Kent, A. G., Vill, A. C., Shi, Q., Satlin, M. J. & Brito, I. L. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 11, 4379 (2020).
Smillie, C. S. et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 (2011).
Groussin, M. et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184, 2053–2067.e18 (2021).
Frazão, N., Sousa, A., Lässig, M. & Gordo, I. Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proc. Natl Acad. Sci. USA 116, 17906–17915 (2019).
Muller, H. J. Some genetic aspects of sex. Am. Nat. 66, 118–138 (1932).
Hill, W. G. & Robertson, A. The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294 (1966).
Liu, Z. & Good, B. H. Dynamics of bacterial recombination in the human gut microbiome. PLoS Biol. 22, e3002472 (2024).
Garud, N. R., Good, B. H., Hallatschek, O. & Pollard, K. S. Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. PLoS Biol. 17, e3000102 (2019).
Ghalayini, M. et al. Evolution of a dominant natural isolate of Escherichia coli in the human gut over the course of a year suggests a neutral evolution with reduced effective population size. Appl. Environ. Microbiol. 84, e02377–17 (2018).
Kersten, K., de Visser, K. E., van Miltenburg, M. H. & Jonkers, J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 9, 137–153 (2017).
AACR Project GENIE Consortium. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 7, 818–831 (2017).
Collisson, E. A. et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Gerstung, M. et al. The evolutionary history of 2,658 cancers. Nature 578, 122–128 (2020).
Shibata, H. et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 278, 120–123 (1997).
Sun, J. et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014).
Xue, W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014).
Platt, R. J. et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
McFadden, D. G. et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 156, 1298–1311 (2014).
Sánchez-Rivera, F. J. et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014).
Nguyen, L. V. et al. Barcoding reveals complex clonal dynamics of de novo transformed human mammary cells. Nature 528, 267–271 (2015).
Bhang, H. C. et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 21, 440–448 (2015).
Rogers, Z. N. et al. A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nat. Methods 14, 737–742 (2017). This work pioneered Tuba-seq, enabling systematic quantification of tumour suppressor gene function in vivo and transforming our ability to study cancer evolution directly in living organisms.
Winters, I. P. et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat. Commun. 8, 2053 (2017).
Cai, H. et al. A functional taxonomy of tumor suppression in oncogenic KRAS-driven lung cancer. Cancer Discov. 11, 1754–1773 (2021).
Foggetti, G. et al. Genetic determinants of EGFR-driven lung cancer growth and therapeutic response in vivo. Cancer Discov. 11, 1736–1753 (2021).
Hebert, J. D. et al. Combinatorial in vivo genome editing identifies widespread epistasis and an accessible fitness landscape during lung tumorigenesis. Mol. Biol. Evol. 42, msaf023 (2025).
Rogers, Z. N. et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat. Genet. 50, 483–486 (2018).
Martino, M. E. et al. Bacterial adaptation to the host’s diet is a key evolutionary force shaping drosophila-lactobacillus symbiosis. Cell Host Microbe 24, 109 (2018).
Lebov, J. F., Schlomann, B. H., Robinson, C. D. & Bohannan, B. J. M. Phenotypic parallelism during experimental adaptation of a free-living bacterium to the zebrafish gut. mBio 11, e01519–e01520 (2020).
Robinson, C. D. et al. Experimental bacterial adaptation to the zebrafish gut reveals a primary role for immigration. PLoS Biol. 16, e2006893 (2018).
Ekroth, A. K. E., Gerth, M., Stevens, E. J., Ford, S. A. & King, K. C. Host genotype and genetic diversity shape the evolution of a novel bacterial infection. ISME J. 15, 2146–2157 (2021).
Barlow, J. T., Bogatyrev, S. R. & Ismagilov, R. F. A quantitative sequencing framework for absolute abundance measurements of mucosal and lumenal microbial communities. Nat. Commun. 11, 2590 (2020).
Giraud, A. et al. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291, 2606–2608 (2001).
Yilmaz, B. et al. Long-term evolution and short-term adaptation of microbiota strains and sub-strains in mice. Cell Host Microbe 29, 650–663.e9 (2021).
Barroso-Batista, J. et al. The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 10, e1004182 (2014).
Barroso-Batista, J., Demengeot, J. & Gordo, I. Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat. Commun. 6, 8945 (2015).
Barreto, H. C., Sousa, A. & Gordo, I. The landscape of adaptive evolution of a gut commensal bacteria in aging mice. Curr. Biol. 30, 1102–1109.e5 (2020).
Lescat, M. et al. Using long-term experimental evolution to uncover the patterns and determinants of molecular evolution of an Escherichia coli natural isolate in the streptomycin-treated mouse gut. Mol. Ecol. 26, 1802–1817 (2017).
Lam, L. H. & Monack, D. M. Intraspecies competition for niches in the distal gut dictate transmission during persistent salmonella infection. PLoS Pathog. 10, e1004527 (2014).
Vasquez, K. S. et al. High-resolution lineage tracking of within-host evolution and strain transmission in a human gut symbiont across ecological scales. Preprint at bioRxiv https://doi.org/10.1101/2024.02.17.580834 (2024).
Vasquez, K. S. et al. Quantifying rapid bacterial evolution and transmission within the mouse intestine. Cell Host Microbe 29, 1454–1468.e4 (2021).
Wong, D. P. G. H. & Good, B. H. Quantifying the adaptive landscape of commensal gut bacteria using high-resolution lineage tracking. Nat. Commun. 15, 1605 (2024).
Grubaugh, N. D. et al. Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 4, 10–19 (2019).
Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018).
Maurano, M. T. et al. Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York city region. Genome Res. 30, 1781–1788 (2020).
Feder, A. F. et al. More effective drugs lead to harder selective sweeps in the evolution of drug resistance in HIV-1. eLife 5, e10670 (2016).
Roark, R. S. et al. Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth. Science 371, eabd2638 (2021).
Dapa, T. et al. Within-host evolution of the gut microbiome. Curr. Opin. Microbiol. 71, 102258 (2023).
Good, B. H. & Rosenfeld, L. B. Eco-evolutionary feedbacks in the human gut microbiome. Nat. Commun. 14, 7146 (2023).
Rego-Costa, A., Huang, I. T., Desai, M. M. & Gombert, A. K. Yeast population dynamics in Brazilian bioethanol production. G3 13, jkad104 (2023).
Large, C. R. L. et al. Genomic stability and adaptation of beer brewing yeasts during serial repitching in the brewery. Preprint at bioRxiv https://doi.org/10.1101/2020.06.26.166157 (2020).
Garge, R. K. et al. Systematic profiling of ale yeast protein dynamics across fermentation and repitching. G3 14, jkad293 (2024).
Bitter, M. C. et al. Continuously fluctuating selection reveals fine granularity of adaptation. Nature 634, 389–396 (2024).
Rudman, S. M. et al. Direct observation of adaptive tracking on ecological time scales in Drosophila. Science 375, eabj7484 (2022).
Bitter, M. C. et al. Pervasive fitness trade-offs revealed by rapid adaptation in large experimental populations of Drosophila melanogaster. Preprint at bioRxiv https://doi.org/10.1101/2024.10.28.620721 (2024).
Biller, S. J., Berube, P. M., Lindell, D. & Chisholm, S. W. Prochlorococcus: the structure and function of collective diversity. Nat. Rev. Microbiol. 13, 13–27 (2015).
Kinsler, G., Geiler-Samerotte, K. & Petrov, D. A. Fitness variation across subtle environmental perturbations reveals local modularity and global pleiotropy of adaptation. eLife 9, e61271 (2020).